http://www.chemistrymag.org/cji/2008/104020pe.htm |
Apr. 1,
2008 Vol.10 No.4 P.20 Copyright |
(College of Chemistry and Environmental Science, Hebei University, Baoding 071002)
Abstract In this paper, a sensitive resonance light scattering (RLS) method for
determination of thrombin by the incorporation of oligonucleotides-functionalized gold
nanoparticles (GNPs) was developed. The oligonucleotide probe 1 and probe 2 were designed
complementary to the part of the DNA aptamer of human a -thrombin respectively and
immobilized on the surface of GNPs. The hybridization between the oligonucleotide probes
on the GNPs and the aptamer can result in the aggregation of the GNPs and great
enhancement of RLS intensity. The specific interaction between the aptamer and thrombin can restrain the hybridization and result
in the decrease of RLS intensity, which is well dependent on the thrombin concentration.
Based on the phenomena, thrombin can be sensitively determined in the concentration range
of 6.0-32.5 nmol/L and
the detection limit (3s) is 4.3 nmol/L.
Keyword gold nanoparticles (GNPs), aptamer, human a-thrombin,
resonance light scattering (RLS)
1.
INTRODUCTION
Thrombin, a multifunctional serine protease, recognizes multiple macro-molecular
substrates and plays a key role in both procoagulant and anticoagulant functions[1].
So the detection of thrombin is very important for clinical diagnosis and biological
research. Aptamers are short oligonucleotides selected in vitro selection process[2]
for specific that can bind to a broad range of molecular targets with high-affinity, such
as amino acids, drugs, proteins and other molecules, and are considered as promising
recognition elements for biosensor applications.
The discovery of aptamer offers a new way for the detection of
thrombin. A series of detection techniques using aptamers to determine thrombin have been
reported, such as fluorescence[3-7], chemiluinescence[8],
colorimetric[9-11], elecreochemistry[12,13], and AFM[14].
Among them, fluorescence detection is most widely used. Unfortunately, most of these
measurement techniques need fluorescence labeling or some other functional changes of the
aptamer before designing the aptamer probes, which may result in high-cost, complicated
designing and operating. In recent years, great attention has been focused on the use of
nanoparticle labels on the aptamer in order to overcome the problems associated with
fluorescent labels and other functional. For example, V. Pavlov, et al. designed a
colorimetric method based on gold nanoparticles to detect thrombin in homogeneous and
heterogeneous forms[11]. Since Pasternack et al.[15] had applied the
resonance light scattering (RLS) technique to study the aggregation of porphyrin and
chlorophyll, this technique gradually drew many researchers' attention because
of high sensitivity and operational simplicity. As a new spectral analysis technique, the
RLS technique has been widely used to quantitatively determine proteins[16,17] in
aqueous solution in view of the enhanced RLS signals.
In the present paper, we report a novel effective approach that utilizes the
oligonucleotide-functionalized gold nanoparticle (GNP) probes and the aptamer coupled with
the RLS technique for quantitatively detecting thrombin in homogeneous solution. The
thiol-capped oligonucleotide probe 1 and probe 2 were designed complementary to part of
the aptamer, respectively. By functionalizing the GNPs with either 3'- or 5'- termini of
the thiol-capped oligonucleotide probes, the hybridization of the aptamer and the probes
will lead oligonucleotide-functionalized GNPs to aggregate to form a network structure,
which results in great enhancement of RLS intensity. In the present of human a-thrombin, aptamer firstly interacts with thrombin, which results in the
suppression of the hybridization between the aptamer and the oligonucleotide probes on the
GNPs and the decrease of RLS intensity. Based on this, we explored a simple and sensitive
RLS method for thrombin determination.
2.
EXPERIMENTAL
2.1 Apparatus
A Hitachi F-4500 fluorescence spectrophotometer (Tokyo, Japan), equipped with a 150 W
high pressure Xenon lamp, was used to measure and record the RLS spectra and RLS
intensity. The absorption spectra were obtained by a TU-1901 UV-Vis spectrophotometer
(Beijing Purkinje General Instrument Co., Ltd, Beijing, China). A GL-16G-high speed
refrigerated centrifuge (Shanghai Anting Scientific Instrument Co., Ltd, Shanghai China)
was used for the centrifugation of Au nanoparticle solution. A WH-861 vortex mixer
(Taicang Instrumental Co., Jiangsu, China) was used to blend the solutions. A thermostatic
water bath (Liaota) was used to control the temperature for the interaction between
proteins and their aptamers.
2.2 Reagents
Tetrachloroauric acid (HAuCl4·4H2O) was obtained from
Sinopharm Group Chemical Reagent Co., Ltd reagent Inc. Human a-thrombin (all of the thrombin mentioned below is human a-thrombin) was obtained from Haemotologic Technologies, Inc. The DNA
thrombin-binding aptamer and two 3'-and 5'-alkanethiol-modified oligonucleotides were
purchased from Takara Biotechnology (Dalian) Co., Ltd. Their sequences were listed as
follows: aptamer: 5'- AGT CCG TGG TAG GGC AGG TTG GGG TGA CT - 3'; probe 1: 5' SH
-TTT TTT TTT TAG TCA CCC CAA CCT - 3'; probe 2: 5'- CCC TAC CAC GGA CTT TTT TTT TTT -SH
3'. All other reagents were of analytical reagent grade and used as purchased without
further purification. Doubly distilled water was used throughout.
The GNP was prepared according to the literature[18]. All
glassware used in the preparation was thoroughly cleaned with chromate washings (cleansing
solution), rinsed in water, and oven-dried prior to use. A 100 mL of 0.01% HAuCl4
solution was heated to boil, and then 3.5 mL of the 1.0% trisodium citrate solution was
quickly added with vigorously stirring. Boiling was continued for another 6 min. The
solution was naturally cooled to room temperature after moving away from the heater, and
then stored at 4 ºC. The
prepared GNPs were characterized by TEM image as about 16 nm diameter[18].
GNPs were
modified with alkanethiol oligonucleotides by adding alkanethiol oligonucleotides to 1 mL
aqueous GNP solution to a final oligonucleotide concentration of 1 mmol/L. After being incubated for 24 h at room temperature, the
solution was adjusted to the pH value and ionic strength of the phosphate buffer (10
mmol/L, pH 7.0) and allowed to stand for 8 h. Then 2 mol/L NaCl was added to the solution
to bring the total NaCl concentration of the solution to 0.05 mol/L. This was repeated 16
h later by adjusting the NaCl concentration to 0.1 mol/L. The solution was allowed to "age" under these conditions for
additional 24 h. Excess oligonucleotides were then removed by centrifugation for 30 min at
12000 rpm. Following removal of the supernatant, the red oily precipitate was washed three
times with 0.1 mol/L NaCl, 10 mmol/L phosphate buffer (pH 7.0) by successive
centrifugation. Finally, the red oily precipitate was redispersed in 1 mL doubly
distilled water and stored at 4 ºC. The
working solution of the oligonucleotode probes-functionalized GNPs was obtained by mixing
the prepared 40 mL oligonucleotode probe 1-functionalized
GNPs and 40 mL oligonucleotode probe 2-functionalized GNPs and
then diluting the mixture to 200 mL with a buffer solution
consisting of 20 mmol/L Tris-HCl (pH 7.4) and 0.3 mol/L NaCl (5-fold dilution).
2.3 Standard procedure
All the tubes that were used for the interaction between aptamer and thrombin were
blocked by 3% BSA for 1h before used. The aptamer at final concentration of 10 nmol/L and
appropriate amount of thrombin were mixted in the Tris-HCl buffer solution (20 mmol/L, pH
7.4). The mixture was incubated at 25 ºC for 20 min. Then 20 mL of the solution was added into 180 mL working solution of oligonucleotide probes-functionalized GNPs.
The solution was blended thoroughly by vortex mixer. After incubation at room temperature
for 3h, RLS spectra were obtained with scanning synchronously with the same excitation and
emission wavelengths from 200 to 700 nm by fluorescence spectrophometer and the RLS
intensities were measured at the wavelength of 550 nm. The slit width and PMT voltage of
the measurements were 5 nm and 400 V, respectively.
3.RESULT AND
DISCUSSION
3.1 Spectral characteristics
As shown in Fig.1, the oligonucleotide-functionalized GNP solution (composed of 180 mL working solution of the oligonucleotide-functionalized GNP
solution and 20 mL Tris-HCl buffer (20 mmol/L, pH 7.4)
exhibits two low light scattering peaks at 300 nm and 550 nm, respectively. The absorption
peaks of 16 nm GNPs appear at 520 nm [11] due to their surface plasmon
oscillation. According to the RLS theory[20], the light scattering peak of the
GNPs at 550 nm can be characterized as RLS peak, which is very near the wavelength of
maximum absorption of the GNPs.
Because the oligonucleotide probe 1 and probe 2 on the GNPs were
complementary to the part of the aptamer respectively, when the GNP solution was mixed
with the aptamer, the sandwich hybridization between the probes and the aptamer would
result in the aggregation of the GNPs with a network structure. One can see from Fig.1c,
the aggregation greatly enhanced the RLS intensity of the GNPs.
On the other
hand, if the aptamer was firstly mixed with thrombin, the specific interaction between the
aptamer and thrombin would prevent the hybridization of the aptamer with the
oligonucleotide probes. Fig.1b showed that the prevention decreased the enhanced RLS
intensity. The decrease of the RLS intensity would be dependent on the amount of thrombin,
based on which the relationship between RLS intensities and the amounts of thrombin could
be established and the amount of thrombin could be sensitively determined.
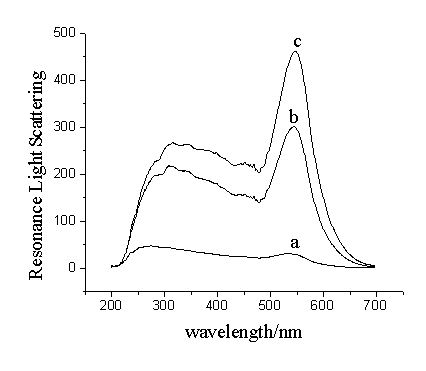
Fig.1 Resonance light scattering spectra
of (a) oligonucleotide-functionalized GNP working solution, (b)
oligonucleotide-functionalized GNP working solution in the presence of 10 nmol/L aptamer
and 30 nmol/L thrombin, (c) oligonucleotide-functionalized GNP working solution in the
presence of 10 nmol/L aptamer
3.2
Optimization of the amounts of aptamer and nucleotide probe- functionalized GNPs
The proposed RLS method for
determination of thrombin was based on the thrombin restraining the hybridization of the
aptamer and the oligonucleotide probes on GNPs and the concentration of thrombin was
determinated through the decrease of RLS intensity. Therefore, the linear range and the
sensitivity for determination of thrombin would be respectively dependent on the dynamic
range and the slope of the relationship between RLS intensities and the aptamer
concentrations. Compared curve a with curve b in Fig.2, it can be seen that the dynamic
range and the slop would be largely increased with increasing the concentration of
oligonucleotide probe-functionalized GNPs when the 10-fold dilution and 5-fold dilution of
the GNP solution were used. The dynamic range and the slope by using the 3-fold dilution
of the GNP solution increased very little than that by using the 5-fold dilution of the
GNP solution (Fig.2c). In addition, the use of 3-fold dilution of the GNP solution would
increase the reagent consumption of the GNPs. Therefore, the 5-fold dilution of the GNP
solution was selected as the working solution for determination of thrombin. On the other
hand, the dynamic range of the aptamer was in the concentration range of 0.5 ~ 10 nmol/L.
Thus, 10 nmol/L aptamer was used as the optimum concentration in order to obtain a widely
linear range for determination of thrombin.
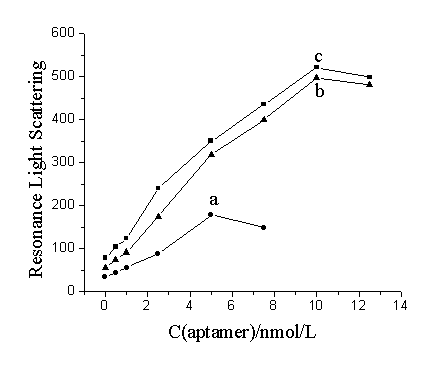
Fig.2 The relationship between RLS
intensities and aptamer concentration. The used solution of oligonucleotide
probes-functionalized GNPs was prepared by mixing 60 mL probe 1-GNP solution and 60 mL probe 2-GNP solution, and
then diluting the mixture to (a) 600 mL (10-fold dilution); (b) 300 mL (5-fold dilution); (c) 180 mL (3-fold
dilution) with Tris-HCl buffer (20 mmol/L, pH 7.4)
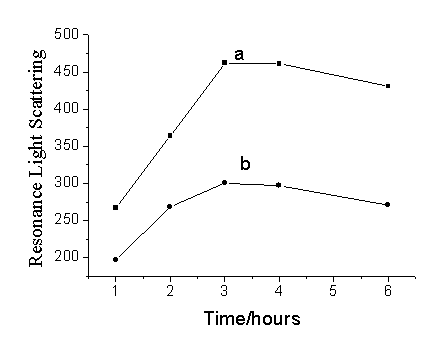
Fig.3 The influence of hybridization time on
the RLS intensity of (a) blank: the working solution of oligonucleotide
probes-functionalized GNPs and 10 nmol/L aptamer; (b) sample: the working solution of
oligonucleotide probes-functionalized GNPs and 10 nmol/L aptamer in the present of 30
nmol/L thrombin.
3.3 Influence of the hybridization time on the RLS intensity
The influence of hybridization time on RLS intensity was investigated. 10 nmol/L aptamer
and 30 nmol/L thrombin were firstly mixed in the Tris-HCl buffer solution (20 mmol/L, pH
7.4), and then incubated at 25 ºC for 20 min.
RLS intensities were measured after mixing 20 mL incubated
solution and 180 mL working solution of the oligonucleotide
probes-functionalized GNPs at different times. The same mixture solution without thrombin
was used as the blank. As shown in Fig.3, both of the RLS intensities of the sample and
blank reached the maximum after incubating 3 hours, kept a constant at least 1 hour, and
then gradually decreased. Therefore, we chose 3 hours as the best detection time when the
RLS intensity ratio of the sample to the blank had a maximum value and kept a constant
after 3 hours.
3.4 Calibration curves
According to the standard procedure, an investigation of the relationship between the
RLS intensity and the concentration of thrombin was carried out under the optimized
experimental conditions mentioned above. As shown in Fig. 4, I0 is the RLS
intensity of the working solution of oligonucleotide probes-functionalized GNPs by
addition of the 10 nmol/L aptamer. I is the RLS intensity after adding thrombin. It is
indicated from the results that there is a good linear relationship between (I0-I)/I0
and thrombin concentrations when thrombin is in the range of 6.0 - 32.5 nmol/L, and the
detection limit (3s) was estimated to be 4.3 nmol/L.
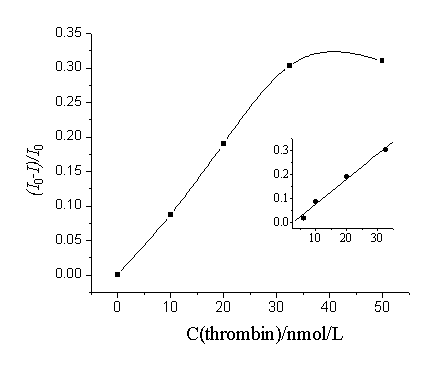
Fig.4 Relationship
between (I0-I)/I0 and the
concentration of thrombin
4. CONCLUSION
From the results mentioned above, we can conclude that the hybridization between the
oligonucleotide probes on GNPs with the aptamer can result in the aggregation of the GNPs
and the great enhancement of RLS intensity of the GNPs. The specific interaction between
the aptamer and thrombin can prevent the hybridization because the aptamer can form
G-quadruplex structure[21] and decrease the RLS intensity. The decrease of the
RLS intensity is well dependent on thrombin concentration. Based on this, thrombin can be
determined with high sensitivity and specificity. As low as nmol/L level thrombin can be
detectable with the proposed method. What's more, since aptamer can recognize the protein
strongly and selectively, it could be considered as functional analogues of monoclonal
antibodies, which has many advantages to antibody (such as their facile in vitro
synthesis, cheep cost, thermal stability, easy modification and high purity). Aptamers
would become an ideal class of molecules for protein analysis. If change to another
aptamer, it's target protein will be detected quickly
and sensitively. Therefore, the RLS method by using aptmers may open up a new possibility
for biological assays and clinical diagnosis.
ACKNOWLEDGEMENT The project was supported by the National Science Foundation of China (NSFC, No. 20675023) and the National Science Foundation of Hebei Province (No. 2006000967).
REFERENCES
[1] Diane M Tasset, Mark F Kubik, Walter Steiner. J. Mol. Biol, 1997, 272: 688.
[2] Ellington A D, Szostak J W. Nature, 1990, 346: 818.
[3] Fang X, Sen A, Vicens, et al. Chem. Biochem, 2003, 4: 829.
[4] Myoyong Lee, David R Wakt. Analytical Biochemistry, 2000, 282: 142.
[5] Jiangwei J Li, Xiaohong Fang, Weihong Tan. Biochemical and Biophysical Research
Communications, 2002, 292: 31.
[6] Nobuko Hamaguchi, Andrew Ellington, Martin Stanon. Analytical Biochemstry, 2001,
294:126.
[7] Milan N Stojanovic, Paloma De Prada, Donald W Landry. J. Am. Chem. Soc. 2001,
123:4928.
[8] Yaxin Jiang, Xiaohong Fang, Chunli Bai. Anal. Chem, 2004, 76: 5230.
[9] Milan N Stojanovic, Donald W Landry. J. Am. Chem. Soc., 2002, 124: 9678.
[10] Chih Ching Huang, Yu Fen Huang, Zehui Cao, et al. Anal. Chem., 2005, 77: 5735.
[11] Valeri Pavlov, Yi Xiao, Bella Shlyahovsky, et al. J. Am. Chem. Soc., 2002, 126:11768.
[12] Kazunori Ikebukuro, Chiharu Kiyohara, Kojo Spde. Biosensors and Bioelectronics, 2005,
20: 2168.
[13] Daniel A Di Giusto, Wjatschesslaw A Wlassoff, J Justin Gooding. Nucleic Acids
Research. 2005, 33: e64.
[14] Jiang Y, Zhu C, Ling L, et al. Anal. Chem., 2003, 75: 2112.
[15] Pasternack R F, Bustamante C, Collings P J et al. J. Am. Chem. Soc.,1993,115: 5393.
[16] Huang C Z, Li Y F, Feng P et al. Fresenius. J. Anal. Chem., 2001,371: 1034.
[17] Jia R P, Zhai H L, Shen Y et al. Talanta, 2004, 64: 355.
[18] Zheng Ping Li, Xin Rui Duan, Cheng Hui Liu et al. Analytical Biochemistry, 2006,
351:18.
[19] K C Grabar, R. G Freeman, M B Hommer, et al. Anal. Chem, 1995, 67: 735.
[20] R F Pasternack, P J Collings. Science, 1995, 269: 935.
[21] Tasset D M, Kublik M F, Steiner W. Journal of Molecular Biology, 1997,272(5): 688.
李正平,李玉佩,李炜
(保定河北大学化学与环境科学学院,071002)
摘要 基于共振光散射检测技术和寡聚核苷酸功能化的金纳米粒子探针,本文发展建立了一种灵敏测定凝血酶的新方法。设计寡聚核苷酸探针1和探针2分别与凝血酶核酸适体的一部分匹配杂交。当用探针1和探针2功能化的金纳米粒子与此核酸适体混合,此探针与核酸适体的杂交导致金纳米粒子的聚集并引起共振光散射的强烈增强;而核酸适体与凝血酶的特异性结合将抑制此探针与核酸适体的杂交并导致共振光散射强度的降低。基于此现象,凝血酶在6.0-32.5 nmol/L范围内可被灵敏地测定,相应的检出限 (3s)为4.3 nmol/L。
关键词 金纳米粒子,核酸适体,凝血酶,共振光散射