|
Molbank 2006, M484 |
Synthesis, Physical
Characterization, Antibacterial and Antifungal activities of a novel bis(3-((E)-1-(2-hydroxyphenyl)ethylideneamino)phenyl)methanone
A. F. Jalbout*1,
A. A. Jarrahpour*2, J. M. Brunel3,
C. Salmi3,
1Department of
Chemistry, the
2Department of Chemistry,
3Laboratoire SESNAB, Facult®¶ de St J®¶rôme,
Case 342, Universit®¶ Paul C®¶zanne, Av. Escadrille Normandie Ni®¶men, 13397 Marseille cedex 20,
France
Phone: +98 711 2284822,
Fax: +98 711 2280926, e-mail:[email protected] or [email protected]
*Author to whom correspondence should be addressed;
Received: 13 January 2006 / Accepted: 3
March 2006 / Published: 1 June 2006
Keywords:
2-Hydroxyacetophenone, 3, 3'- diaminobenzophenone,
Schiff base, AM1 and B3LYP/6-31G*
Abstract: Bis (3-((E)-1-(2-hydroxyphenyl) ethylideneamino) phenyl) methanone has been synthesized in this paper and its
structure was confirmed by 1H-NMR, 13C-NMR, IR and Mass
spectra. Its AM1 and B3LYP/6-31G*
calculations to characterize the physical properties of this molecule has been
also presented. Finally, the antifungal and antibacterial activities of
this derivative have been evaluated.
Introduction
Particular
attention has recently been paid to the synthesis and study of the diimino tetradentate Schiff bases
and their complexes. This is due to their uses as biological models in
understanding the structure of biomolecules and
biological processes. The crucial role of Schiff bases in the biological
function of bacteriorhodospin is proven. The retinal chromophore is bound covalently to the protein via a protonated Schiff base [1].Schiff-base ligands derived from 2-hydroxyacetophenone have been used
as complexing agents [2]. In these ligands the electronic, steric
and geometric effect of a methyl group on an imine
carbon on asymmetric catalytic reactions can be investigated [3]. Salen type complexes are
very well studied because this type of Schiff base ligands
present suitable biometric properties that can mimic the structural feature of
the active site[4].Based on these facts we decided to
synthesize a new bis Schiff base derived from 2-hydroxyacetophenone.
Results and Discussion:
3,3'-Diaminobenzophenone 1 (0.212g, 1.00mmol) and 2-Hydroxyacetophenone 2 (0.272g, 2.00mmol) were dissolved in 10 mL of warm ethanol. The reaction mixture was refluxed for 10h and allowed to stand aside. The crystals were filtered off and washed with ethanol. The pure Schiff base 3 was recrystalized from ethanol as light yellow crystals. The IR spectrum showed the characteristic absorption of Schiff base C=N at 1612 cm-1. The 1H-NMR spectrum showed a multiplet for aromatic protons at 6.76-7.85 ppm. The OH group appeared as a singlet at 14.37 ppm. The 13C-NMR spectrum showed C=N group at 162.23, C=O group at 196.97 and the CH3 group at (18.48, 30.61). The mass spectrum showed the (M+1) peak at 449. The yield was 57%.
Melting point: 182-184 °„C.
IR (KBr, cm-1): 3248; 1716; 1612 cm-1.
1H NMR (250 MHz, CDCl3): ¶ń= 14.37 (OH); 7.85- 6.76 (Aromatic protons); 1.65 (s, CH3).
13C NMR (62.9 MHz, CDCl3): ¶ń= 196.97; 162.23; 148.62; 138.29; 117.67; 116.98; 114.34; 30.61; 18.48.
MS: 449 (M+1), 195, 120, 92, 65, 43.
All calculations in this work where carried out with the AM1 level of
theory using the GAUSSIAN 03 [5] suite of programs. In addition we have carried
very intense B3LYP/6-31G* optimizations and frequency calculations. More
information about these methods is available elsewhere [6]. Figure 1 presents
the optimized structure of the molecule with bond lengths and bond angles shown
as well as the theoretical IR vibrational spectrum.
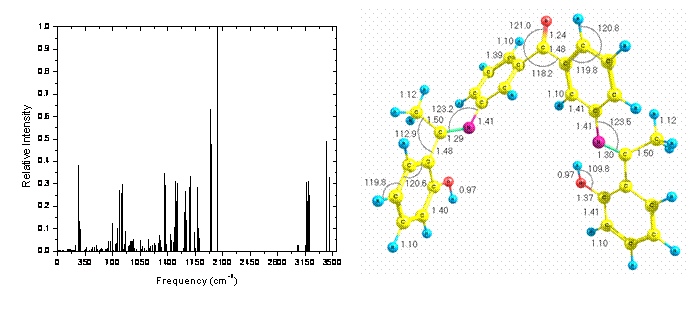
Figure 1. AM1 optimized structure and its theoretical IR vibrational spectrum for molecule 3.
|
|
Fitted Thermodynamic
Equation (T/1000=t) |
100 K |
298.15 K |
1000 K
|
|
Cp |
61.26809+ 1473.5495*t -123.39616*t2 -341.74085*t3 -0.0486*t-2 |
205.78 |
477.77 |
1077.52 |
|
S |
53.05624*ln(t) + 1474.23257*t + 423.72982*t2/2 -484.29762*t3/3
-32.45372/(2*t2) -3357.13221 |
518.49 |
863.88 |
1807.1 |
|
H |
89.57261*t + 1689.9106*t2/2 -308.96521*t3/3
-332.76221*t4/4 + 0.78161/t -370.051 |
13.13 |
80.56 |
670.35 |
Table 1: Thermodynamic properties of the molecule 3 in Figure 1, calculated at the AM1 level and B3LYP/6-31G* level of theory, where Cp is the heat capacity in J mol-1 K-1, S is the entropy in J mol-1 K-1, and ¶§H is the standard enthalpy kJ mol-1. These where fitted to the Shomate equations which are implemented by the JANAF tables of the NIST databases. These equations converged to an R2 value of 0.999 on average.
Table 1 shows the thermodynamic properties for the
where T (temperature in K), S (entropy in J mol-1 K-1), Cp (heat molecule in
Figure 1 capacity at constant pressure in kJ mol-1 K-1),
and ¶§H=H°„ - H°„298.15 (enthalpy content, in kJ mol-1), T1=100 K,
T2=298.15 K, and T3=1000 K calculated AM1 and B3LYP/6-31G* frequencies. The
fits were performed according to the equations implemented by the National
Institute of Standards and Technology (NIST) [7]. These equations have been
very good at predicting physical properties of various molecules, as we have
tested in the past [6-10].
Antibacterial
and antifungal tests
Derivative 3 was evaluated for its in vitro biological properties
against human pathogens [11].
This compound was found to possess no antifungal activities against S. cerevisiae (ATCC 28383) and no
antibacterial activities against Gram-positive and Gram-negative bacteria even
if a slight antibacterial activity against S. aureus
(CIP 4.83) at a
concentration of 100 µg/mL
has been noticed (Table 2).
|
Sample CIP |
Antimicrobial activity (MIC), µg/mL |
|||
S. cerevisiae
(ATCC 28383) |
S. aureus (4.83) |
C. albicans (1180-79) |
E. Coli (54127) |
|
|
3 |
>100 |
100 |
>100 |
>100 |
Table 2: Antimicrobial activity of Schiff
base 3
Acknowledgment
AFJ and BT would like to
thank the
References
1. Boghaei, D.M., Gharagozlou M., Spectrochimica Acta Part A 61. 2005, 3061®C3065.
2. a) Zhang, J. X.; Zhou, Y.; Cai, G. J. Mol. Catal, 1997,
11, 41-44; (b) Holland, D; Laidler, D.A.;
Milner, D.J. J. Mol. Catal, 1981, 11,119-127. (c)
Chen, G. M.; Chen, F.;
Zhou, C, Chem. Chin. Univ, 1995, 16, 216-219. (d) Qiu,
M.; Liu, G.; Yiao, X, Chin. J.Catal,
2001, 22, 77-80. (e) Li, C.; Zhang, W.;
3. Gao, W. T.; Zheng, Z.; Molecules 2002, 7, 511-516.
4. Karmakar, R., Choudhury, C. R., Bravic, G., Sutter, J. P., Mitra, S., Polyhedron, 23 2004, 949®C954.
5. Frisch,
M.J., et.al., GAUSSIAN 03, Revision A.1, M. J. Frisch, et. Al., Gaussian, Inc.,
6. Foresman,
J.B., Æ Frisch, , Exploring Chemistry with Electronic Structure Methods, 2nd
edition Gaussian, INC, Pittsburgh, PA, 1996
7. Linstrom,
P.J., Mallard, W.G., Eds., NIST Chemistry WebBook,
NIST Standard Reference Database Number 69, July 2001, National
Institute of Standards and Technology, Gaithersburg MD, 20899 (http://webbook.nist.gov).
8. Jalbout,
A.F. , Solimannejad, M., Labonowski, J.K., Chem. Phys. Lett.,
2003, 379, 503.
9. Jalbout,
A.F., Jiang, Z.-Y., Quasri, A., Jeghnou,
H., Rhandour, A., Dhamelincourt,
M.C., Dhamelincourt, P., Mazzah,
A., Vib. Spect.,
2003, 33, 21.
10. Jalbout,
A.F., Nazara, F., Turker,
L., J. Mol. Struct. (THEOCHEM), 2004,
627, 1. (Invited Review)
11. Growth was measured in
vitro using a liquid-phase method according to NCCLS guidelines from the
American Society of Microbiology for 24 hours using various concentrations of drugs.Dei Cas, E.; Dujardin, L.; Ribeiro Pinto, M.
E.; Ajana, F.; Fruit, J.; Poulain,
D.; Camus, D.; Vernes, A.; Francois, N. Mycoses 1991, 34, 167-172
Sample Availability: Available from MDPI.
© 2006 MDPI. All rights reserved.