http://www.chemistrymag.org/cji/2001/039046ne.htm |
Sep. 1,
2001 Vol.3 No.9 P.46 Copyright |
Synthesis and crystal structure of Cu(I) mixed-ligand complex [Cu(dpa)(PPh3)2](BF4)· CH3CN· CHCl3
Li Dan, Li Rongzhen, Qi Zhiyu, Shi Xuhua,
Feng Xiaolong#, Cai Jiwen#
(Department of Chemistry, Shantou University, Shantou, Guangdong 515063; #Instrumentation Analysis & Research Center, Zhongshan
University, Guangzhou, 510275, China)
Received Mar.2, 2001; Supported by the National Natural Science Foundation of China (No.29901004), Natural Science Foundation of Guangdong Province of China(No. 000779) and project 211 of Shantou University.
Abstract Reaction of [Cu(CH3CN)2(PPh3)2](BF4)
with 2,2'-dipyridylamine (dpa) afforded the mononuclear copper(I) mixed-ligand complex,
[Cu(dpa)(PPh3)2](BF4)· CH3CN ·CHCl3.
The complex was characterized with elemental analysis, infrared spectrometry and X-ray
crystallography. The complex cation consisted of a distorted tetrahedral N2P2 core,
contributed by triphenylphosphine and 2,2'-dipyridylamine,
respectively.
Keywords copper(I) complex, triphenylphosphine, 2,2'-dipyridylamine, crystal
structure
In the previous papers, the copper(I)-phosphine coordination chemistry have been reported[4-6]. As an extension of these studies we now introduce a bidentate heterocyclic nitrogen ligand, 2,2'-dipyridylamine (dpa) to form a copper(I) mixed-ligand complex. The synthesis and crystal structure of [Cu(dpa)(PPh3)2](BF4)· CH3CN · CHCl3 are presented.
1 EXPERIMENTAL
1.1 Instruments and materials
CHN elemental analyses were performed by using a Perkin-Elmer 240C analyser. Infrared
absorption spectra were recorded on a Nicolet Magna 750 FT-IR spectrometer with KBr
tablets. Triphenylphosphine (Merk), 2,2'-dipyridylamine (Aldrich) and other reagents for
synthesis are commercially available without further purification.
1.2 Preparation
To a chloroform solution (10 ml) of [Cu(CH3CN)2(PPh3)2](BF4)
[7] (0.76g, 1 mmol) was added 2,2'-dipyridylamine (0.17g, 1 mmol). The light
yellow solution was stirred 4 hours and turn to light green. Keep stirring for another 7
hours, the resulting solution was then filtered and concentrated to ca. 5 ml. The
solid product was obtained by addition of absolute diethyl ether to the solution.
Recrystallization of the crude product from a chloroform-n-hexane solution afforded
a colorless crystal that is suitable for X-ray crystallographic determination. The
crystals were identified as [Cu(dpa)(PPh3)2](BF4)· CH3CN
·CHCl3 (Elemental analysis for the crystal sample: Found C, 58.41; H, 4.21; N,
5.65%. Calc. for C49H43BCl3CuF4N4P2:
C, 58.47; H, 4.31; N, 5.57%).
1.3 Crystallography
A single crystal with dimensions of 0.24mm×0.12mm×0.11mm was selected to collect the diffraction intensity data on a
Brucker SMART CCD diffractometer fitted with graphite-monochromated Mo Ka radiation, l= 0.071073 nm. The measurement was carried out at room temperature.
All of the calculations were performed by using the SHELXTL package. The structure was
solved by direct methods and refined by full matrix least squares. The total of 23702
reflections were collected in the range of 4.03°<q<24.71°. The unique reflections are 8296 with Rint
= 0.0357. Goodness of fit on F2 is 1.055. Final R indices [I>2s (I)] R1 = 0.0722, wR2
= 0.2172. Largest diff. Peak and hole are 1.174 and 0.743 e·Å-3,
respectively.
2 RESULTS AND DISCUSSION
2.1 Synthesis and infrared spectrum
Title complex was obtained from the ligand substitution reaction of [Cu(CH3CN)2(PPh3)2](BF4).
Two CH3CN ligands were replaced by dpa, containing two pyridine nitrogen atoms
which coordinated to copper. Although most copper(I) compounds may be oxidized easily in
air, the title complex is stable. This may be due to the coordination of copper with the
reducing ligand triphenylphosphine.
The IR spectrum of the title complex shows a serial of strong peaks at
492, 504, 517, 695, 744, 1027, 1434, 1479 cm-1 from PPh3. The broad
band at around 3450 cm-1 and very strong peaks at 1579, 1631 cm-1
are due to N-H and C=N stretching vibration of dpa, respectively, and several weak bands
at 2852, 2922 and 3053 cm-1 can be assigned to the aliphatic C-H stretching
vibration. A strong broad band at 1084 cm-1 indicates the existence of BF4-
ion.
2.2 Crystal structure
The empirical formula of the title complex is C49H43BCl3CuF4N4P2,
formula weight 1006.51. The crystal is monoclinic space group P21/n with a
= 14.530(4) Å, b =
18.954(5) Å, c = 18.244(5) Å, a= 90° , b = 102.960(4)° , g = 90° , V = 4896(2) Å3, Z = 4 and Dc
= 1.365 mg/m3, m
= 0.728 mm-1, F(000) = 2064. Non-hydrogen coordinates and thermal parameters
are listed in Table 1. Selected bond lengths and angles are given in Table 2. Figure 1 and
2 show the structure and unit-cell of the title complex, respectively.
Table 1 Non-Hydrogen Atomic Coordinates (×104) and Thermal Parameters (Å2×103)
|
y |
|
U(eq) |
|
Cu(1) |
6460(1) |
805(1) |
3097(1) |
45(1) |
Table 2 Selected bond lengths and angles of the title complex
bond |
length/Å | angle |
|
| Cu(1)-N(1) Cu(1)-N(3) Cu(1)-P(1) Cu(1)-P(2) |
2.070(4) 2.087(4) 2.2859(15) 2.2649(15) |
N(1)-Cu(1)-N(3) |
91.18(18) |
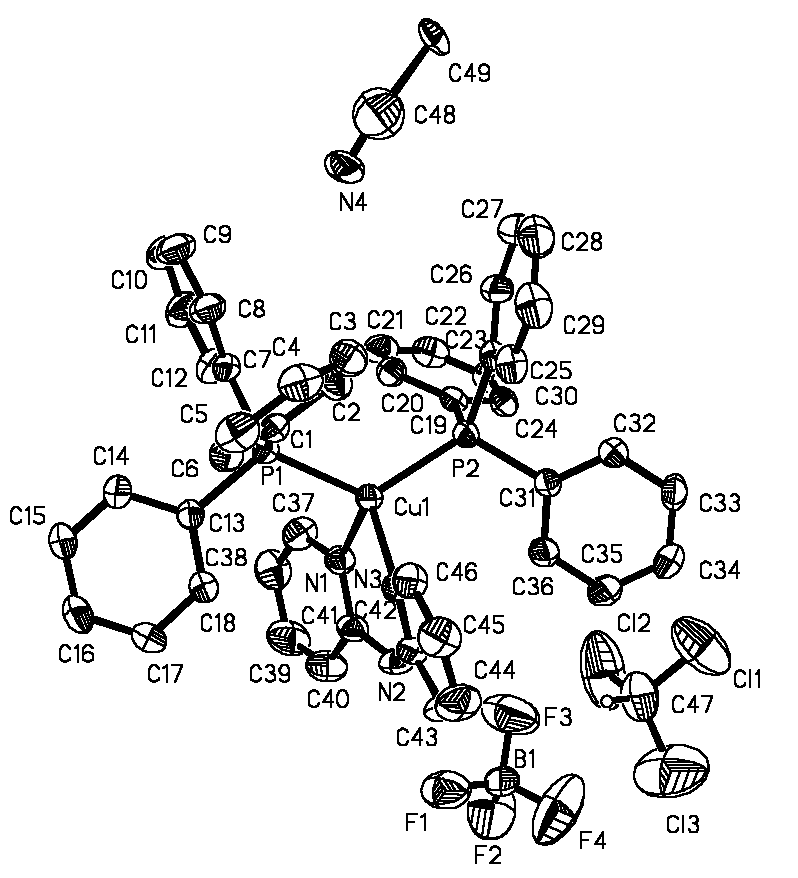
Figure 1 Molecular structure of [Cu(dpa)(PPh3)2](BF4)· CH3CN ·CHCl3 with atom numbering schemes
Table 3 Selected bond lengths and angles of related copper(I) complexes
| complex cation | Cu-P (Å) |
Cu-N (Å) |
P-Cu-P (°) |
N-Cu-N (°) |
ref. |
[Cu(phen)(PPh3)2]+ |
2.271(1) |
2.070(2) |
115.44(4) |
80.90(9) |
8 |
[Cu(bipy)(PPh3)2]+ |
2.246(3) |
2.056(8) |
125.4(1) |
79.6(4) |
9 |
[Cu(py)2(PPh3)2]+ |
2.271(4) |
2.102(7) |
115.85(9) |
101.5(2) |
9 |
[Cu(dpa)(PPh3)2]+ |
2.2859(15) |
2.070(4) |
124.99(5) |
91.18(18) |
this |
As shown in Figure 1, title
complex is mononuclear Cu(I) complex. The structure consists of discrete [Cu(dpa)(PPh3)2]+
and BF4- ions, with CH3CN and CHCl3 solvent
molecules. There is no special reciprocity between two adjacent molecules (Figure 2). In
[Cu(dpa)(PPh3)2]+ cation, the coordination geometry about
Cu(I) can be described as distorted tetrahedral with a N2P2 donor set, contributed by PPh3
and dpa. For comparison, Table 3 summarized selected bond lengths and angles of several
similar Cu(I) complexes with CuN2P2 core. In [Cu(dpa)(PPh3)2]+,
the Cu-N distances of 2.070(4) and 2.087(4) Å are comparable to those of complexes listed
in Table 3, but are much longer than those in Cu(II) complex [Cu(dpa)(PPr)2] 2H2O
(HPPr = phenylpropanoic acid) with the values of 1.979(3) and 1.975(3) Å. [10]
The Cu(I) complexes in Table 3 have similar Cu-P distances. The P-Cu-P and N-Cu-N angles
of 124.99(5) and 91.18(18)° are quite different from those in [Cu(py)2(PPh3)2]+
(115.85(9) and 101.5(2)°), indicating the angle distortions are much greater in
[Cu(dpa)(PPh3)2]+ than in pseudotetrahedral [Cu(py)2(PPh3)2]+.
Obviously, this is due to that two coordination sites are occupied by a bidentate ligand
dpa.
The crystal structure of title complex has been deposited at the
Cambridge Crystallographic Data Centre with the deposition number of CCDC 165706.
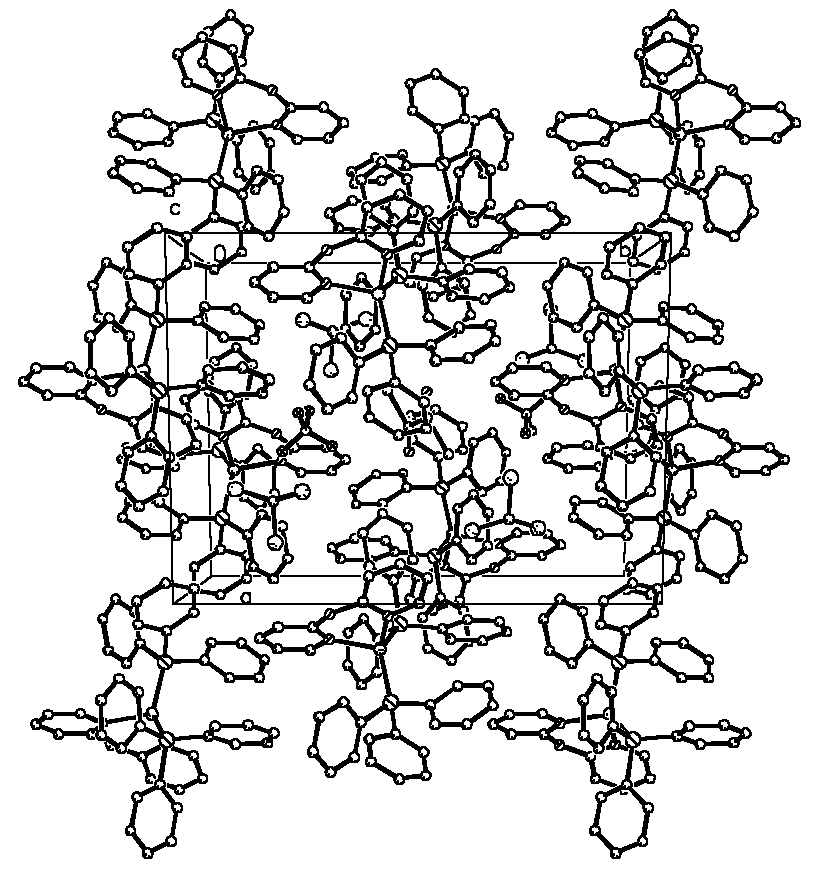
Figure 2 Unit-cell contents of [Cu(dpa)(PPh3)2](BF4)· CH3CN ·CHCl3
REFERENCES
[1] Karlin K D, Zubieta J (eds.). Copper Co-ordination Chemistry Biochemical and
Inorganic Perspectives, New York: Adenine Guilderlend, 1983.
[2] Kitajima N, Moro-oka. Chem. Rev., 1994, 94: 737.
[3] Karlin K D, Tyeklar Z (eds.). Bioinorganic Chemistry of Copper, New York: Chapman
& Hall, 1993.
[4] Li Dan, Yip H K, Che C M et al. J. Chem. Soc., Dalton Trans, 1992, 2445.
[5] Li Dan, Che C M, Wong W T et al. J. Chem. Soc., Dalton Trans, 1993, 653.
[6] Li Dan, Yang K E, Huang S W, Jiang Q Z. Spectroscopy and Spectral Analysis, 1996, 16:
10.
[7] Diez J, Falagan S, Gamasa P et al. Polyhedron, 1988, 7 (1): 37.
[8] Kirchhoff J R, McMillin D R, Robinson W R et al. Inorg. Chem., 1985, 24 (23): 3928.
[9] Engelhardt L M, Pakawatchi C, White A H, Healy P C. J. Chem. Soc., Dalton Trans, 1985,
125.
[10] Feng X W, Gong Y Q, Jiang Y T et al. Chinese J. Inorg. Chem., 1999, 15 (2): 243.