http://www.chemistrymag.org/cji/2003/05a082pe.htm |
Oct. 1, 2003 Vol.5 No.10 P.82 Copyright |
Determination of nucleic acids by resonance light scattering technique with victoria blue B
Chen Zhanguang, Liao Xuhong, Li Dan, Ding Weifeng(Department of Chemistry, Shantou University, Shantou 515063, China) Abstract This is the first report of the sensitizing effect of Cetyltrimethylammonium bromide (CTMAB) on the enhanced resonance light scattering (RLS) resulted from the interaction of organic dye Victoria blue B (VBB) with nucleic acids. In the buffer solution (pH 6.80), the interactions of VBB with nucleic acids gave two characteristic peaks of RLS at 390.0 and 444.0 nm. The interaction was sensitized by the presence of CTMAB. Under the optimum conditions, the enhanced RLS intensity at 390.0 nm was proportional to the concentrations of nucleic acids in the range 0.05 - 2.0mg mL-1 for fsDNA , ctDNA, and 0.02 - 1.5mg mL-1 for yRNA. The detection limits (3s) were 5.61, 7.22, 3.03 ng mL-1 for fsDNA, ctDNA and yRNA, respectively. Synthetic samples were determined with good reproducibility.
Keywords Cetyltrimethylammonium bromide; Nucleic acids; Victoria blue B; resonance light scattering
1 INTRODUCTION
Victoria blue B (VBB) is an easily available triphenylmethane dye. Although VBB can be bound to the polyanion of DNA through chromophore coupling between VBB and DNA bases owing to the positive charge on its molecular structure, the RLS intensity cannot be enhanced significantly. In our experiment, we found that the interactions between VBB and nucleic acids can be sensitized by the presence of CTMAB. Based on this phenomenon, a novel assay of nucleic acids was established. In this paper, the nature of resonance light scattering of the ion-association complex of VBB-CTMAB - nucleic acids system and the mechanism of the reaction have been studied for the first time. The relationship between DIRLS and nucleic acid concentration was established. Synthetic samples have been determined by this assay, and the results are reproducible and reliable.
2 EXPERIMENTAL
2.1 Reagents
The stock solutions of nucleic acids were prepared by dissolving commercial
products in doubly distilled water. The concentration of the working solution was 25 mg mL-1.
The nucleic acids used in this study are fsDNA (Sigma Chemicals Co., USA), ctDNA
(Sino-American Biotechnology Co., China), and yRNA (Sigma Chemicals Co., USA). These
stocks needed to be stored at 0¨C4ºC.
The absorbances ratios A260/A280 were 1.83 (DNA) and
2.0 (RNA).
A 8.0¡Á10-6 mol L-1 working solution of VBB was
prepared by dissolving commercial product (Shanghai Chemical Reagent Co., Shanghai, China)
in 10% ethanol.
A 8.0¡Á10-4 mol L-1 working solution of
surfactant CTMAB was prepared by dissolving its crystal product (Shanghai Chemical Reagent
Co.) in doubly distilled water.
NaAc buffer was prepared by dissolving sodium acetate in doubly
distilled water, and the pH value was adjusted to 6.80 with NaOH. Its working solution was
2.0 mol L-1. All chemicals used were of analytical grade or the best grade
commercially available and doubly distilled water was used throughout.
2.2 Apparatus
The RLS spectra and intensities were measured with a Perkin-Elmer LS55
luminescence spectrometer with a quartz cuvette (1¡Á1 cm). All absorbance measurements
were measured on a Shimadzu UV-2501 ultraviolet spectrophotometer (Kyoto, Japan). A SA 720
laboratory instrument (Orion Research) was used to measure the pH value of the solution.
2.3 General Procedure
Into a 10ml standard flask were added 1.5 mL
of NaAc buffer, 0.5 ml of VBB, 0.4 ml of CTMAB, and appropriate standard nucleic acids or
sample solution. The mixture was mixed thoroughly after each addition of the interacting
reagents, and then diluted to the mark with doubly distilled water. 10 min's incubation
time was needed.
RLS spectra were obtained by scanning simultaneously the excitation and
emission monochromators of the LS55 spectrofluorometer from 250.0 to 600.0 nm with Dl= 0 nm. Both
the excitation and emission slit widths were kept as 10.0 nm. The RLS intensities were
measured at 390.0 nm.
3 RESULTS AND DISSUSIONS
3.1 Features of RLS spectra
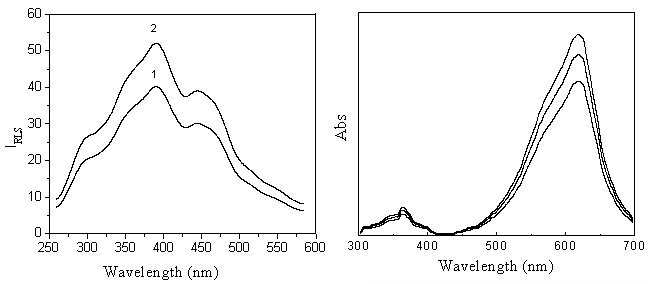
Fig.1 displays the RLS spectra of fsDNA and VBB respectively. It can be seen that VBB has
two wide weak RLS peak located at 390 and 430 nm. Although VBB exists mainly as R+
cationic monomer in the medium of pH 6.80, and nucleic acid exists as a big polyvalent
anionic, the RLS intensity of VBB was just enhanced a little by the addition of fsDNA
(Fig.1).
As Fig.2 shows, VBB has two absorption bands characterized at 370 and
615 nm. The absorption intensity increases with the increasing of VBB concentration. This
indicates that VBB has aggregate tendency in the reaction medium. According to the RLS
theory [1,2,10], the RLS
peak at 390 nm is ascribed to the absorption band at 370 nm. The weaker RLS in the
wavelength range > 500 nm, however, is ascribed to the molecular absorption band of VBB
over 500-700 nm on one hand, and to the steep decrease of light scattering intensity with
wavelength increase on the other.
 |
|
| Fig.1 RLS spectra of the interaction of VBB and fsDNA Concentrations: curve 1: VBB 4.0¡Á10-7 mol L-1; curve 2: fsDNA 1.0mg mL-1, VBB 4.0 ¡Á10-7 mol L-1; pH, 6.80 | Fig. 2 Absorption spectra of VBB. Concentrations: VBB, (from top to bottom, ¡Á10-7 mol L-1) 4.0, 3.0 2.0; pH, 6.80 |
3.2 Optimal conditions of the interaction
3.2.1 Effect of CTMAB
Strong RLS signals can be observed when CTMAB is added to the reaction
system. Fig. 3 displays the role that CTMAB played in the interactive system. As can be
seen, CTMAB has little effect on the RLS intensity of VBB because of the electrostatic
repulsion. This indicates that the presence of CTMAB sensitized the reaction. The RLS
intensity of VBB - CTMAB - fsDNA increases with the CTMAB concentration increasing. The
optimum CTMAB concentration is in the range 3.0 ¡Á 10-5 - 3.7 ¡Á 10-5
mol L-1, and when the concentration of CTMAB is greater than 3.7 ¡Á 10-5
mol L-1, the formation of micelle resulted in the decrease of RLS intensity.
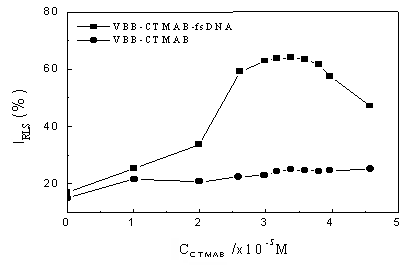
Fig. 3 Dependence of RLS intensity on CTMAB concentration. Conditions:
VBB, 4.0 ¡Á10-7 mol L-1; fsDNA, 1.0mg mL-1; pH, 6.80
3.2.2 Effect of pH and buffers
The dependence of RLS intensity on pH values of the interaction medium has been studied.
The optimum pH range is from 6.68 to 6.90. Any pH value out of the range leads to a lower
RLS intensity. Thus, pH 6.80 is always used for the interactive medium. Experiments
indicate that the different buffers have different effects on the RLS intensity. The
following buffers were tested: Tris - HCl, KH2PO4 - HCl, NaH2PO4
- NaOH and NaAc - NaOH. The results show that NaAc is the best buffer and the
optimum volume of buffer is 1.5 mL. In this work, the optimal addition order of the
reagents is as follows: buffer solution -VBB-CTMAB-nucleic acids.
3.2.2 Effect of VBB concentration
VBB concentration also has significant effects on the interactive system. RLS intensity
increases when VBB concentration is less than 4.0¡Á10-7 mol L-1, and
decreases when VBB concentration is greater than 4.0¡Á10-7 mol L-1.
So we selected 4.0¡Á10-7 mol L-1 as the optimum VBB concentration
for determination of nucleic acids in this method.
3.2.3 Effect of ion strength
We used NaCl to control the ion strength of the reaction system. The result showed that
the enhanced RLS intensity is very sensitive to the change of ionic strength. When the ion
strength was lower than 0.015 mol L-1, the RLS intensity had no significant
change. But when the ion strength was higher than 0.015 mol L-1, the RLS
intensity increase and the reaction system became unstable. So, in our experiment, we need
to control the ion strength at a low level.
3.3 Tolerance of foreign substances
The effects of foreign substances such as metal ions, proteins, sugars,
nucleotides and surfactants on the RLS intensity of the interactive system were tested.
The results are listed in Table 1. It can be seen that proteins, sugar and nucleotides do
not interfere the DNA. The common metal ions
in biological fluids, such as Na+, Ca2+, Mg2+ and K+
can be tolerated at high concentration. However, easily hydrated metal ions, such as
Fe(III), Al (II), Cu(II) in near-neutral solution, could form Fe(OH)3, Al(OH)3
and Cu(OH)2 particles respectively, and result in light scattering, thus they
have a positive effect on the light scattering intensity of the studied system. Therefore,
these metal ions have lower tolerances.
Substances |
Concentration |
Change of |
Substances |
Concentration |
Change of |
Cu(II), SO42- |
1.0 |
5.5 |
L-Alanine |
1.0 |
- 1.5 |
Al(III), SO42- |
1.2 |
7.6 |
Urea | 100 |
4.7 |
Ca(II), Cl- |
30 |
7.2 |
SLS | 2.0 | - 2.5 |
Co(II), Cl- |
1.3 |
- 2.8 |
Tween 80 | 0.02a | 1.7 |
Zn(II), SO42- |
5.0 |
- 6.2 |
Triton X-100 | 0.02a |
3.6 |
K(I), SO42- |
30 |
5.9 |
Protein, BSA | 2.0b | 4.9 |
Mg(II), Cl- |
20 |
3.2 |
5'-ADP | 1.0 | 3.4 |
Fe(III), Cl- |
1.0 |
- 5.6 |
5'-GDP | 1.0 |
- 2.5 |
Mn(II), SO42- |
0.8 |
- 5.8 |
5'-TDP | 1.0 |
- 1.6 |
Pb(II),Cl- |
1.6 |
5'-CDP | 1.0 |
2.3 | |
Glucose |
5.0 |
2.3 |
4. Analytical applications
4.1. Calibration and detection limit
Under the optimum conditions according to above the standard procedure,
good linear relationships were obtained between the enhanced RLS intensity at 390.0 nm and
the concentration of nucleic acids. All the regression parameters are presented in Table
2. The sensitivities of the RLS method for different nucleic acids have the sequence:
fsDNA£¾yRNA£¾ctDNA.
Nucleic acid |
Linear range ( mg mL-1) |
Regression equation |
Detection limit |
Correlation coefficient (r) |
fsDNA |
0.05-2.0 |
D IRLS = - 1.43 + 43.4 c |
5.61 |
0.9984 |
ctDNA |
0.05-2.0 |
D IRLS = 3.31 + 38.2 c |
7.22 |
0.9951 |
yRNA |
0.02-1.5 |
D IRLS = 1.54 + 40.5 c |
3.03 |
0.9981 |
Table 3 Recovery ratios of fsDNA in the synthetic samples
Nucleic acids in Sample ( mg mL-1) |
Main additivesa |
Found value |
Recovery range |
RSDb (%) |
fsDNA 1.0 |
Ca(II), Mg(II), BSA |
1.01 |
94.1 - 102.3 |
1.19 |
ctDNA 1.0 |
ADP, GDP, CDP, TDP |
0.98 |
96.3 - 104.8 |
1.25 |
yRNA 1.0 |
Triton X-100, ADP, Fe(II) |
0.99 |
92.1 - 100.2 |
2.55 |
b Relative standard deviation for 5 measurements. Table 4 Comparison of detection limits of nucleic acids between this method and other methods
Methods |
Nucleic acids |
LOD (ng mL-1) |
Reference |
Ethdium Bromide |
(n)DNA |
10 |
11 |
Hoechst 33258 |
(n)DNA |
10 |
12 |
Safranine T (ST) |
fsDNA/ctDNA/yRNA |
39.8/19.8/61.1 |
5 |
Azur A |
fsDNA/ctDNA |
12.6/10.9 |
6 |
| Nile blue sulfate | fsDNA/ctDNA/yRNA |
1.2/0.4/10.5 |
7 |
Morin-CTMAB |
ssDNA/ctDNA/yRNA |
27.0/16.7/23.4 |
13 |
This method |
fsDNA/ctDNA/yRNA |
5.61/7.22/3.03 |
4.2. Determination of trace amounts
of nucleic acids in synthetic samples
Under optimal conditions according to the standard procedures, three
synthetic samples, in which a series of foreign substances had been added based on the
tolerance levels of foreign substances displayed in Table 1, were determined according to
the linear relationship given in Table 2. As Table 3 shows, the recovery ratios of the
synthetic samples range from 92.1% to 104.8%, and the RSD is less than 2.55%. The results
for the synthetic samples indicate that this assay is sensitive, reproducible and
reliable.
A comparison between this method and other common methods for nucleic
acids in sensitivity is summed up in Table 4. It can be seen that the sensitivity of this
method is better than most of well-known methods.
REFERENCES
[1] Pasternack R F, Bustamante C, Collings P J et al. J. Am Chem Soc.,
1993, 115: 5393.
[2] Pasternack R F, Collings P J. Science, 1995, 269: 935.
[3] Huang C Z, Li K A, Tong S Y. Anal. Chem., 1996, 68: 2259.
[4] Huang C Z, Li K A, Tong S Y. Anal. Chem., 1997, 69: 514.
[5] Huang C Z, Li Y F, Liu X D. Anal. Chim. Acta, 1998, 375: 89.
[6] Li Y F, Huang C Z, Huang X H, et al. Anal. Chim. Acta, 2001, 429: 311.
[7] Huang C Z, Li Y F, Li N B et al. J. Anal. Chem. (Fenxi Huaxue), 1999, 27: 1241.
[8] Liu Y, Ma C Q, Li K A et al. Anal. Chim. Acta, 1999, 379: 39.
[9] Gibbs E J, Tinnoco Jr. I, Maestre M F et al. Biochem. Biophys. Res. Commun., 1988,
157: 350.
[10] Miller G A. J Phys. Chem., 1978, 82 (5): 616.
[11] Lepeeq J B, Paoletti C. Anal. Biochem., 1966, 17: 100.
[12] Labara C, Paigen K. Anal. Biochem., 1980, 102: 344.
[13] Liu R T, Yang J H, Wu X et al. Anal. Chim. Acta, 2001, 448: 85.