http://www.chemistrymag.org/cji/2005/074032pe.htm |
Apr.28, 2005 Vol.7 No.4 P.32 Copyright |
(Dept. of Chemistry, Jinan University, Guangzhou 510632, China; #Guangzhou Jinan Biomedicine Research & Development Base, Guangzhou 510632, China) Received Jan. 31, 2005; Supported by National Natural Science Foundation of China (No.20271022), the Key Project of Education Ministry of China (01141), and Guangzhou Natural Science and Technology Project (2001-J-010-01). Abstract A reversed phase high performance liquid chromatographic (RP-HPLC) method for simultaneous determination of 19 amino acids in selenium-enriched Spirulina platensis (Se-SP) was established. The samples were derivatized with dansyl (Dns-Cl) and analyzed on a C18 column at 30oC, using a binary gradient elution with a fluorescent detector (lex: 320 nm; lem: 523 nm). The peaks area of the 19 amino acids linearly correlated with their concentrations. Detection limit was 3.5×10-8 mol/L. The quantitative analysis showed that Se-enriched treatment did not change the content of the common amino acids in Se-SP. The content of Selenomethionine (Se-Met) was 0.261 mg/g in the Se-SP. The detection linearity, precision and recovery associated with this assay were evaluated. It was the first time to use an HPLC method to simultaneously determine SeMet and the common amino acids.
Keywords HPLC; Selenomethionine; Amino acid; Selenium; Spirulina platensis. The trace mineral selenium (Se) is an essential nutrient element of fundamental importance to human biology. This has become increasingly obvious as new researches have shown a hitherto unsuspected role for this element to human health[1]. Se lowering may cause such diseases as cardiomyopathy, cancer, endemic osteoarthropathy, anemia, etc [2]. Our previous works have shown that Spirulina platensis (S. platensis) can be cultured under high Se(IV) concentrations and transform the inorganic Se to organic Se at a high rate [2-5], which indicate that S. platensis was good carrier for Se biotransformation. It is well known that the bioavailability and the toxicity of Se compounds are closely correlated with their chemical forms. Information on speciation of Se in natural products is vital. For these reasons the demands for accurate and sensitive methods for Se speciation in nutritional supplements, e.g. Se-enriched garlic, yeast, and lactic acid bacteria, have rapidly increased [6]. Selenomethionine (SeMet) was one of the most important seleno amino acids incorporated into proteins of organisms. In vitro and in vivo studies have shown that SeMet exhibited potent anticancer activity in models of cancers of the colon and prostate [7]. In addition, SeMet has been shown to function as the precursor in enzyme-mediated production of methylselenol [8]. The popular and reliable methods for the determination of SeMet were MIP-MS, ICP-MS and GC-ICP-MS[9]. They were all too expensive to be widely used. However, precolumn derivatization of free amino acids with dansyl (Dns-Cl) is economical and can be successfully used for quantitation of amino acids and polyamines over a wide range of relative concentrations of dansyl chloride[10]. It was the first time to use an HPLC method to simultaneously determine SeMet and the common amino acids within a single run in this work, which would save not only time but also costs of solvents and supplies, and waste disposal.
1. EXPERIMENTAL
1.1 Chemicals and instruments
The simultaneous separation of dansylated SeMet and other amino acids was carried out
on an isocratic RP-HPLC (HP1100) system with a C18 column (LiChrospher®100RP-18,
endcapped 5 mm).
1.2 Culture of Se-enriched S. platensis
The cultivation of S. platensis was carried out in 250-mL Erlenmeyer flasks containing 100 mL Zarrouk medium (pH 9.0) at 30oC with a light illumination of 4000 lx and a 14:10 h light:dark cycle. Se was added with the form of Sodium Selenite: Na2SeO3 on the 5th (300mg/L), 6th (400mg/L), 7th (300mg/L) day respectively with an accumulative concentration of 1000 mg/L. Cells were harvested on the 10th day.
1.3 Sample preparation and derivatization
Approximately 100 mg dried mass of Se-enriched S. platensis was placed in 10mL HCl 4moL/L. After removing the dissolved oxygen with nitrogen airflow, the mixture was incubated at 50oC in a water bath for 50 h. After the centrifugation at 6000 rpm for 10 min at 4oC, the supernatant was adjusted to pH 5-6 with Li2CO3-HCl buffer solution (pH=9.50). Then, 20 mL aliquot of sample extracts or amino acids standards were placed in microfuge tubes and vortexed. 100 mL of Dns-Cl (3 mg/mL in acetonitrile) were then added to each tube. The tubes were capped, vortexed and incubated in a water bath at 50oC. After 40 minutes, 20 mL methylammmonium hydrochloric acid was added to the mixture to react with excess Dns-Cl. Following an additional 5 minute incubation, 20mL of the solution was injected into the HPLC system for analysis. The blank runs were conducted using dansylated methylammmonium hydrochloric acid.
1.4 HPLC conditions
Dansylated common amino acids and SeMet were separated simultaneously within a single run. The details of the gradient profile used in the present study are described below: (T, B% ): 0,14; 7, 20; 14, 26.5; 20, 27; 22, 28; 35, 68; 45, 68; 46, 14. The mobile phase was consisted of A (10mM NaH2PO4 buffer (pH 6.55) containing 4% DMFM and B (100% acetonitrile: ACN). The column heater was set at 30oC. The excitation and emission wavelengths were set at 320 and 523 nm, respectively. The HPLC flow rate was 1.0 mL / min.
1.5 Peak detection, Recovery, Precision and Detection limit
The peak quantifications were done by external standard method using five-point calibration curves. The sample matrices were spiked with known amounts of the common amino acids and SeMet. Blank samples were also prepared in the sample matrices by adding equivalent amounts of water. The recovery of the analyte in each matrix was determined by measuring the ratio of the peak area of analyte to external standard using the standard curves. The analytical precision of this method was evaluated by replicated analysis of the samples. The detection limit was defined based on a signal-to-noise ratio of 3. A new calibration standard set was prepared and analyzed each day. The recoveries were determined in triplicate.
1.6 Determination of total Se(IV), inorganic Se and organic Se concentrations
Se concentrations were determined by using a modified diaminonaphthalene (DAN) fluorescent method [11] with 970CRT fluorescent spectrophotometer, where lex=377nm, lem =523 nm. The dried mass of Se-enriched S. platensis was resuspended in 5 ml of mixture of perchloric and concentrated nitric acid and then digested in an intelligent digestion system for 6h (180oC). It was then reconstituted to 10 ml using HCl 0.1mol/ L and used for total Se(IV) determination. Inorganic Se content was determined directly using extraction solution of algal powder in 15% HCl. Organic Se could be calculated from the content difference between total Se and the inorganic Se.
2. RESULTS AND DISCUSSION
2.1 Optimization of the procedure
The pH value of the mobile phase was an important factor affecting the separation of SeMet
from the common amino acids. We have found the most effective reaction pH to be 6.55 at 30
2.2 HPLC analysis of dansylated amino acids standards
Dansylated amino acids, including SeMet, were separated within a single run using a modification of the HPLC gradient profile described in the previous report [12]. In detail, it was (T, B% ): 0,14; 7, 20; 14, 26.5; 20, 27; 22, 28; 35, 68; 45, 68; 46, 14 with a cleaning method for 30 minutes before shutting down the system. Typical chromatograms of dansylated samples are shown in Fig. 1. The retention times of the analytes were present in Table 1. Peaks were identified by retention time and their fluorescent emission spectra. The emission spectrum of dansylated SeMet was shown in Fig.2, which was quite different from other dansylated amino acids. As shown in Fig 1(1), blank run showed obvious peaks at 7.02, 38.03, 44.96 min. The dansylated amino acids could be separated and detected within a single run. None of the peaks interfered with the others. The separation was completed within 50 minutes.
In the optimized HPLC conditions, the detection limit of this mothed was 3.5×10-8 mol/L. The linear range of detection for amino acids was 0.01-2.5 mmol/mL in most cases, with exceptions of 0.01-4 mmol/mL for SeMet and 0.01-1.25 mmol/mL for Cys (CysH) as listed in Talbe 1. The r2 values for standard curves were 0.991 or higher for all amino acids except for Lys of 0.978. The recovery of the analyte in each matrix was determined by measuring the ratio of the peak area of analyte to external standard using the standard curves. As a result, good recovery of the detected amino acids ranging from 85% to 108% was obtained. The relative standard deviations (RSDs) of the amino acids were less than 5.0 % in most cases, showing a high reproducibility and a good stability for the chromatographic system.
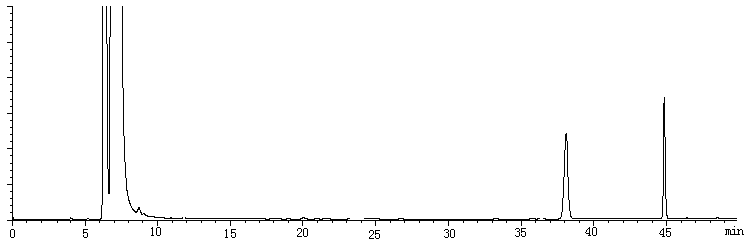
(A)
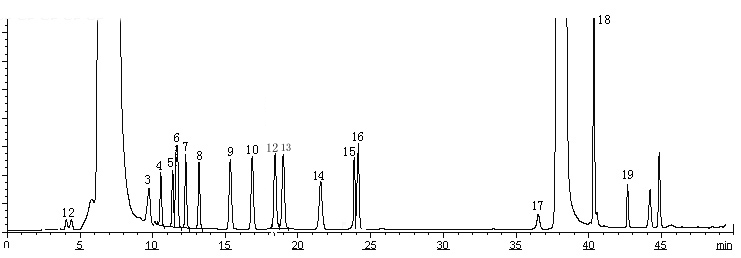
(B)
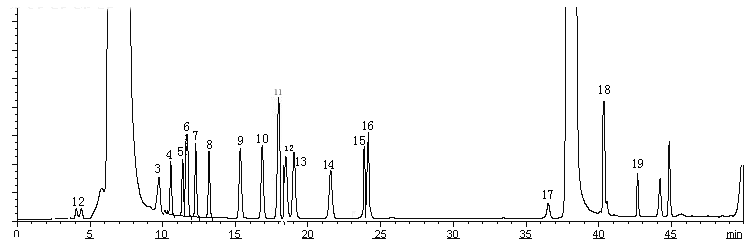
(C)
Fig. 1 Typical chromatograms. (A) Blank run of Dns-Cl. (B) Separation profile of dansyl derivation of 18 amino acids (C) Separation profile of dansyl derivation of 18 amino acids and SeMet.
1 Asp; 2 Glu; 3 Ser; 4 Arg; 5 Thr; 6 Gly; 7 Ala; 8 Pro; 9 Val; 10 Met; 11 Se-Met; 12 Ile; 13 Leu; 14 Phe; 15 Cys; 16 CysH; 17 Lys; 18 His; 19 Tyr.
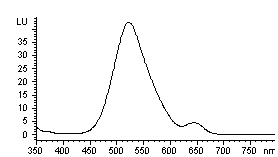
Fig 2 Fluorescent emission spectrum of dansylated SeMet 2.3 HPLC analysis of Se-enriched S. platensis
This method was applied to analyze the amino acids including SeMet in the hydrolyzed samples of Se-enriched S. platensis. As shown in Fig. 3, under the optimized HPLC conditions, a good separation of SeMet from other amino acids was obtained. However, the S. platensis sample contained a large number of unidentifiable dansylated polyamines presenting in the extracts. The contents of the investigated amino acids in Se-enriched S. platensis were shown in Table 1.
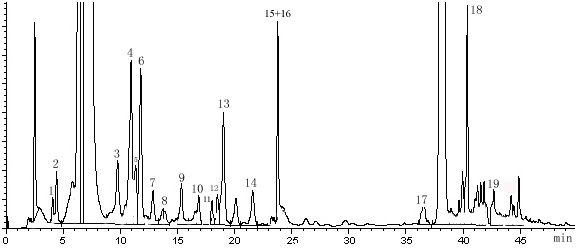
Fig. 3 Chromatograms of dansyl derivation of amino acids in Se-enriched S. platensis
2.3
Total Se(IV), inorganic Se and organic Se content in Se-enriched S. platensisThe results in our study showed that the high Se-enriched S. platensis with total Se content of 1.176 mg/g and organic Se content of 1.024 mg/g could be obtained while the Se was added on the 5th, 6th, 7th day respectively with a final Se concentration of 1000mg/L. The SeMet content was 0.261 mg/g, being equal to 0.0973 mg/g organic Se, which accounted for 9.5% of the total organic Se in Se-enriched S. platensis. The result indicated that S. platensis could bioaccumulate Se efficiently and it was a good carrier for Se enrichment. In addition, except for SeMet, there are many organic Se forms in the Se-enriched S. platensis. The speciation of Se in organisms was calling for the further investigations.
Table 1 Detection limits and linear range for quantification of dansyl amino acids in Se-enriched S. platensis
Amino acids |
Retention time(tR/min) |
Linear range |
Regression equation# |
r2 |
Content in |
RSD |
Asp |
4.071 |
0.01-2.50 |
A=31.172 C-0.929 |
0.998 |
54.12 ± 2.14 |
3.95 |
Glu |
4.405 |
0.01-2.50 |
A=31.055 C-0.662 |
0.998 |
81.43 ± 2.78 |
3.41 |
Ser |
9.752 |
0.01-2.50 |
A=123.84 C+2.705 |
0.999 |
23.71 ± 1.09 |
4.59 |
Arg |
10.764 |
0.01-2.50 |
A=167.48 C+1.205 |
0.999 |
28.17 ± 1.02 |
3.62 |
Thr |
10.951 |
0.01-2.50 |
A=168.51 C-1.135 |
0.999 |
32.88 ± 2.23 |
6.78 |
Gly |
11.342 |
0.01-2.50 |
A=164.41 C+4.076 |
0.998 |
23.63 ± 1.14 |
4.82 |
Ala |
11.792 |
0.01-2.50 |
A=163.32 C+6.572 |
0.998 |
30.49 ± 0.92 |
3.02 |
Pro |
12.858 |
0.01-2.5 |
A=171.34 C-0.091 |
0.999 |
17.12 ± 0.83 |
4.85 |
Val |
15.339 |
0.01-2.5 |
A=169.72 C+3.608 |
0.997 |
20.81 ± 1.64 |
7.88 |
Met |
16.855 |
0.01-2.5 |
A=169.57 C+3.952 |
0.998 |
9.56 ± 0.33 |
3.45 |
SeMet |
18.174 |
0.01-4.0 |
A =1930.4 C- 42.105 |
0.991 |
0.26 ± 0.02 |
7.69 |
Ile |
18.475 |
0.01-2.5 |
A=169.83 C+3.373 |
0.999 |
20.50 ± 0.57 |
2.78 |
Leu |
19.036 |
0.01-2.5 |
A=168.56 C+6.271 |
0.997 |
32.70 ± 1.23 |
3.76 |
Phe |
21.55 |
0.01-2.5 |
A=170.11C+2.749 |
0.999 |
18.87 ± 1.03 |
5.45 |
Cys+CysH |
473.9 |
0.01-1.25 |
A=467.59 C+20.315 |
0.994 |
11.26 ± 0.74 |
6.57 |
Lys |
36.517 |
0.01-2.5 |
A=40.014 C+4.936 |
0.978 |
19.82 ± 0.21 |
1.06 |
His |
40.338 |
0.01-2.5 |
A=515.85 C-7.039 |
0.998 |
5.90 ± 0.18 |
3.05 |
Tyr |
42.665 |
0.01-2.5 |
A=99.987 C+5.013 |
0.996 |
13.21 ± 0.26 |
1.97 |
## Data were expressed as mean ± SD of three cultures (n = 3).
REFERENCES
[1] Zheng W J, Ouyang Z. 1th Edi, Guangzhou: Jinan University Press, 2001.
[2] Chen C, Zhang P, Hu X et al. Biochem. Biophys. Acta, 1999, 14 (27) : 205–215.
[3] Huang Z, Xiang J J, Zheng W J et al. Communication of Plant Physiology (Zhiwu
Shenglixue Tongxun), 2001, 37 (1): 12-14.
[4] Zheng W J, He H Z, Huang Z et al. Marine Science (Haiyang Kexue), 2003, 27 (10):
73-78.
[5] Huang Z, Zheng W J, Xiang J J, et al. Marine Science (Haiyang Kexue), 2002, 26 (5):
60-62.
[6] Chassaigne H, Chery C C, Bordin G et al. Journal of Chromatography A, 2002, 976:
409-422.
[7] Menter DG, Sabichi AL, Lippman SM. Selenium effects on prostate cell growth. Cancer
Epidemiol, Biomarkers Prev 2000;9 (11): 1171–1182.
[8] Wang Z, Jiang C, Lu J. Induction of caspase-mediated apoptosis and cell-cycle G1
arrest by selenium metabolite methylselenol. Mol Carcinogen 2002; 34 (3): 113-120.
[9] Chatterjee A, Shibata Y, Morita M. Microchemical Journal, 2001, 69: 179-187.
[10] Rakesh M, Stephanie L. Journal of Chromatography A, 2004,1035: 63–73.
[11] Zheng W J, Yu J, Yang F et al. Ecologic Science (Shengtai Kexue), 2003, 22 (4):
305-309.
[12] Ding Y Y, Xie M X, Deng Z W. Journal of Beijing Normal University (Beijing Shifan
Daxue Xuebao), 2001, 37 (4): 526-529.
陈填烽,郑文杰,罗勇#,杨芳,白燕
(暨南大学化学系,广东广州 510632;#广州暨南生物医药研究开发基地,广东广州 510632)
摘要 本文首次报道利用反相高效液相色谱同时测定富硒螺旋藻中的硒代蛋氨酸(Se-Met)及18种通用氨基酸。以丹磺酰氯(Dns-Cl)为柱前衍生剂,用反相C18色谱柱在柱温30oC下采用二元梯度洗脱,用荧光检测器 (lex: 320 nm; lem: 523 nm) 对氨基酸的衍生物进行检测。用外标法进行定量分析。样品处理采用了改进的低温低酸、反复通氮除氧方法。结果显示,Se-Met及18种通用氨基酸的峰面积与其浓度在一定范围内呈良好的线性关系;检出限为3.5×10-8 mol/L;该方法的准确度和精密度良好;富硒处理对钝顶螺旋藻(Spirulina platensis, S. platensis)中各种通用氨基酸含量影响不大;富硒螺旋藻干粉中Se-Met含量约0.261 mg/g,占藻体中有机硒的9.5%。
关键词 高效液相色谱;硒代蛋氨酸;氨基酸;硒;螺旋藻.