| http://www.chemistrymag.org/cji/2006/089058pe.htm | Sep. 8,
2006 Vol.8 No.9 P.58 Copyright |
Predication of boiling points of saturated aliphatic aldehydes and alkanones by artificial neural network
Cheng Qian, Zhang Hongyi
(College of Chemistry and Environmental Science, Hebei University, Baoding 071002, China)
Abstract A 3-6-1 BPNN
(back-propagation neural network) was used to link the relation between molecular
structures and boiling points. The data sets were composed of 73 saturated aliphatic
aldehydes and alkanones, with experimental boiling points ranging from 253.7 K to 631.2 K,
and the number of carbon atoms ranging from 1 to 20. Each compound was characterized by
three parameters obtained directly from its molecular structure information, namely
functional group position index (P), carbon atom numbers (N) and methyl numbers (Nm),
and the three parameters were together selected as inputs to the BPNN constructed in this
paper. There was a good linear relationship between the predicted and experimental boiling
points with regression coefficient of 0.9992, and the root mean square (RMS) error for the
predicted boiling points was within 2.9 K. Through a comparison with multiple linear
regression of experimental boiling points against functional group position index (P),
carbon atom numbers (N) and methyl numbers (Nm), we found that the predicted
boiling points by BPNN were superior than those predicted by multiple linear regression,
which gave 11.6 K for the root mean square (RMS) error for the predicted boiling points.
Compared with topological indices methods reported previously, the 3-6-1 BPNN proposed did
not need additional knowledge or software package to calculate the complicated
descriptors, and the number of selected parameters used for inputs was only 3 which was
the smallest in all of the corresponding reports.
Keywords Quantitative structure-property relationship, Artificial Neural Network,
Boiling point, Aliphatic aldehydes, Aliphatic alkanones.
1. INTRODUCTION
One of the most important purposes of applying mathematical, statistical and
computer-based methods in chemistry is to gain the maximum information about the
properties selected compounds by analyzing the chemical data. As a result, the interest in
quantitative structure-property relationship (QSPR) studies has been increased
substantially in these years.
Although topological index as an important QSPR tool has been
successfully used for prediction of all kinds of physical chemistry properties of organic
compounds, including boiling points, reports about the prediction of boiling points for
saturated aliphatic aldehydes and alkanones are still few, due to the presence of oxygen
in their molecules resulting in more complicated calculation of their topological indices
than that of saturated aliphatic chain hydrocarbons. Balaban et al. [1]
predicted 200 carbonyl compounds contained 127 mono-and dialdehydes and –ketones and 73 esters using five topological indices. Toropov and
Toropova [2] obtained one-variable models of the normal boiling points of
carbonyl compounds by the nearest neighboring code (NNC). Toropov and co-workers [3] have
used simplified molecular input line entry system (SMILES) to model normal boiling points
of acyclic carbonyl compounds. Several works about boiling point of saturated aliphatic
aldehydes and alkanones prediction have also been done using molecule (or atom)
topological index by Lin Zhihua and co-workers [4], Chen Yan [5],
Zhang Xiuli and co-workers [6, 7], Feng Changjun, Yang Weihua [8],
Wang Keqiang [9], and so on. However, the common shortcoming in these reports
is that these models needs many topological indexes obtained through many difficult
calculation steps. Therefore, it is necessary to find a new way for predicting the boiling
points of saturated aliphatic aldehydes and alkanones.
As a powerful chemometric technique, artificial neural network (ANNs)
has been used in the study of QSPR [10, 11]. Among the numerous network
architectures, the popular type for QSPR studies is the multilayer feed-forward network
with back-propagation algorithm, usually called back-propagation neural network (BPNN). As
far as we know, no BPNN method for predicting boiling points of saturated aliphatic
aldehydes and alkanones was reported. In this paper, a 3-6-1 BPNN was used to link the
relation between molecular structures and boiling points. The data sets were composed of
73 saturated aliphatic aldehydes and alkanones, with experimental boiling points ranging
from 253.7 K to 631.2 K, and the number of carbon atoms ranging from 1 to 20. Each
compound was characterized by three parameters obtained directly from its molecular
structure information, namely functional group position index (P), carbon atom numbers (N)
and methyl numbers (Nm), and the three parameters were together selected as
inputs to the BPNN constructed in this paper.
The results obtained by the 3-6-1 BPNN were validated, tested and
compared with the results obtained either in previous reports or by multiple linear
regressions.
2. EXPERIMENTAL METHOD AND DATA
2.1 Theory of artificial neural network (ANN)
ANN models are formed by organizing a large number of simple processing elements (PE),
also called neuron nodes into a sequence of layers and linking these layers with
modifiable weighted interconnection [12]. The schematic representation of
neuron node structure is shown in Fig. 1.
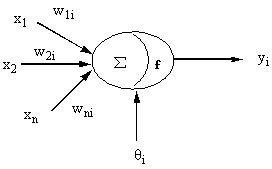
Fig. 1 An artificial neuron model, wni
is the weight associated with the connection from node n to node i, xn
is the output of node i, n is the number of the output, yi is the output
of the node, and
The network input for a node i is given by:
![]()
where j represents nodes in the previous layer, wji is the weight
associated with the connection from node j to node i, xj is the
output of node j, n is the number of the output, and qi is a bias term or threshold value of node i
responsible for accommodating nonzero offsets in the data. The output of node i is
determined by the transfer function and the net input of the node. It is given by:
![]()
where yi is the output of the node i, and
f( ) is the transfer function which we can choose.
The BPNN was used in our
work. The structure of this kind of neural network is shown in Fig. 2. The first layer is
the input layer with one node for each variable or feature of the data. The last layer is
the output layer consisting of one node for each variable to be investigated. In between
there are a series of one or more hidden layer(s). A node in hidden layer(s) can receive
data from any nodes of the anterior layer, process the data, and output a signal.
Fig. 2 A fully connected multilayer feedforward back-propagation network
In the BPNN, processing
obeys back-propagation learning method. The BPNN output and its desired value are
calculated in each iteration process. The changes in the value of weights can be obtained
using the following equation:
![]()
where DWij is the change in the weight factor for each network
node,
The weight plays an important role in the propagation and back-propagation. Actually, a proper setting of these weight factors is essential. The process of adapting the weights to an optimum set of values is called training the neural network. Signals are propagated from the input layer through the hidden layer(s) to the output layer. If the difference between the desired solution and the one obtained does not attain minimum, signals will propagate backward to the input layer. In this course, the weights are adjusting ceaselessly. Through a number of iteration, the difference will reach to minimum. Then, training could be ended.
The back-propagation algorithm has a main problem that it often required a long training course. As a result, there are many modifications to this algorithm. Levenberg-Marquardt (L-M) algorithm is one of the modifications, and it is the learning rule used in our study.
2.2 Software
All calculations were performed on a Founder Feiyue 6000A workstation with 256 MB DRAM memory. The operating system of the Founder PC is Windows XP home edition.
2.3 The data and neural network model
The data set of 73 saturated aliphatic aldehydes and alkanones were taken from the literatures [6,14] and the relationship of their boiling points against their molecular structure information was studied by means of back-propagation neural networks. For each compound, its name and corresponding experimental boiling point (Tb,exp) were given in Table 1 together with three parameters used as inputs in this study, namely the number of carbon atom (N), the carbonyl position index (P) and the number of methyl (Nm). In the three parameters N, P and Nm, the meaning for N and Nm is clear from their names. The carbonyl position index P is defined as the reciprocal of the sequence number for the carbonyl carbon in IUPAC system. Taking 2-methyl butyraldehyde as an example, the first step is to number each carbon in the main chain as IUPAC system. The second step is to find the sequence number for carbonyl carbon. For this example molecule, the sequence number for carbonyl carbon is 1, so its corresponding P is 1. Similarly, if the sequence number for carbonyl carbon was 2 or 3, the corresponding P would be 0.5 and 0.33, respectively. N, P, Nm were the input of the network, and Tb,exp was used as target of the network.
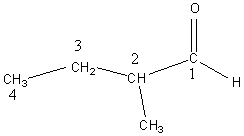
The data set of 73 saturated aliphatic aldehydes and alkanones were randomly divided into two sets: a training set (including 50 groups of data) and a testing set (including 23 groups of data). The compounds collected in the training set and testing set were shown in Table 2 as their serial number in Table 1.
Table1 Data of the 73 saturated aliphatic aldehydes and alkanones, the input and the target of the network.
No. |
Compounds |
Carbon atom numbers |
Carbonyl position index |
Methyl numbers |
Tb,exp/K |
1 |
formaldehyde |
1 |
1 |
0 |
253.7 |
2 |
acetaldehyde |
2 |
1 |
1 |
293.6 |
3 |
propionaldehyde |
3 |
1 |
1 |
321.1 |
4 |
propanone |
3 |
0.5 |
2 |
329.4 |
5 |
n-butyraldehyde |
4 |
1 |
1 |
348 |
6 |
isobutyraldehyde |
4 |
1 |
2 |
337.3 |
7 |
methyl ethyl ketone |
4 |
0.5 |
2 |
352.8 |
8 |
pentanal |
5 |
1 |
1 |
376.2 |
9 |
2-methyl butyraldehyde |
5 |
1 |
2 |
365 |
10 |
3-methyl butyraldehyde |
5 |
1 |
2 |
365.8 |
11 |
2,2-dimethyl propanal |
5 |
1 |
3 |
347 |
12 |
methyl n-propyl ketone |
5 |
0.5 |
2 |
375.5 |
13 |
3-pentanone |
5 |
0.33 |
2 |
375.1 |
14 |
methyl isopropyl ketone |
5 |
0.5 |
3 |
367.4 |
15 |
hexanal |
6 |
1 |
1 |
401.5 |
16 |
2-methyl pentanal |
6 |
1 |
2 |
390.2 |
17 |
3-methyl pentanal |
6 |
1 |
2 |
395.2 |
18 |
4-methyl pentanal |
6 |
1 |
2 |
395.2 |
19 |
2,2-dimethyl butyraldehyde |
6 |
1 |
3 |
377.2 |
20 |
2,3-dimethyl butyraldehyde |
6 |
1 |
3 |
386.2 |
21 |
3,3-dimethyl butyraldehyde |
6 |
1 |
3 |
380.2 |
22 |
2-ethyl butyraldehyde |
6 |
1 |
2 |
390 |
23 |
2-hexanone |
6 |
0.5 |
2 |
400.9 |
24 |
3-hexanone |
6 |
0.33 |
2 |
396.7 |
25 |
3-methyl-2-pentanone |
6 |
0.5 |
3 |
390.6 |
26 |
methyl isobutyl ketone |
6 |
0.5 |
3 |
389.6 |
27 |
2-methyl-3-pentanone |
6 |
0.33 |
3 |
386.6 |
28 |
3,3-dimethyl-2-butanone |
6 |
0.5 |
4 |
379.5 |
29 |
heptaldehyde |
7 |
1 |
1 |
426 |
30 |
3-methyl hexanal |
7 |
1 |
2 |
415.7 |
31 |
2-heptanone |
7 |
0.5 |
2 |
424.1 |
32 |
3-heptanone |
7 |
0.33 |
2 |
420.6 |
33 |
4-heptanone |
7 |
0.25 |
2 |
417.2 |
34 |
3-methyl-2-hexanone |
7 |
0.5 |
3 |
413 |
35 |
4-methyl-2-hexanone |
7 |
0.5 |
3 |
412 |
36 |
5-methyl-2-hexanone |
7 |
0.5 |
3 |
418 |
37 |
2-methyl-3-hexanone |
7 |
0.33 |
3 |
406 |
38 |
4-methyl-3-hexanone |
7 |
0.33 |
3 |
409 |
39 |
5-methyl-3-hexanone |
7 |
0.33 |
3 |
409 |
40 |
3,3-dimethyl-2-pentanone |
7 |
0.5 |
4 |
403.8 |
41 |
3,4-dimethyl-2-pentanone |
7 |
0.5 |
4 |
405 |
42 |
4,4-dimethyl-2-pentanone |
7 |
0.5 |
4 |
398 |
43 |
3-ethyl-2-pentanone |
7 |
0.5 |
3 |
411 |
44 |
2,2-dimethyl-3-pentanone |
7 |
0.33 |
4 |
398 |
45 |
2,4-dimethyl-3-pentanone |
7 |
0.33 |
4 |
397.6 |
46 |
caprylaldehyde |
8 |
1 |
1 |
447 |
47 |
2-ethyl-hexanal |
8 |
1 |
2 |
436 |
48 |
2-octanone |
8 |
0.5 |
2 |
445.8 |
49 |
nonanal |
9 |
1 |
1 |
463.7 |
50 |
3-methyl-2-heptanone |
8 |
0.5 |
3 |
440.2 |
51 |
6-methyl-2-heptanone |
8 |
0.5 |
3 |
440.2 |
52 |
2-methyl-3-heptanone |
8 |
0.33 |
3 |
431.2 |
53 |
6-methyl-3-heptanone |
8 |
0.33 |
3 |
436.2 |
54 |
2-methyl-4-heptanone |
8 |
0.25 |
3 |
428.2 |
55 |
3-ethyl-4-methyl-2-pentanone |
8 |
0.5 |
4 |
427.7 |
56 |
2-nonanone |
9 |
0.5 |
2 |
467.5 |
57 |
5-nonanone |
9 |
0.2 |
2 |
461.6 |
58 |
2 ,6-dimethyl-4-heptanone |
9 |
0.25 |
4 |
441 |
59 |
capraldehyde |
10 |
1 |
1 |
481.7 |
60 |
2-decanone |
10 |
0.5 |
2 |
483.7 |
61 |
3-decanone |
10 |
0.33 |
2 |
476.2 |
62 |
4-decanone |
10 |
0.25 |
2 |
479.7 |
63 |
2-hendecanone | 11 |
0.5 |
2 |
501.2 |
64 |
dodecanal |
12 |
1 |
1 |
522.2 |
65 |
2-decanone |
12 |
0.5 |
2 |
522.2 |
66 |
tetradecanal |
14 |
1 |
1 |
554.2 |
67 |
2-tetradecanone |
14 |
0.5 |
2 |
554.2 |
68 |
hexadecanal |
16 |
1 |
1 |
583.2 |
69 |
2-hexadecanone |
16 |
0.5 |
2 |
583.2 |
70 |
octadecanal |
18 |
1 |
1 |
608.2 |
71 |
2-octadecanone |
18 |
0.5 |
2 |
608.2 |
72 |
eicosanal |
20 |
1 |
1 |
631.2 |
73 |
2-eicosanone |
20 |
0.5 |
2 |
631.2 |
All data were standardized by using the
following equation:
![]()
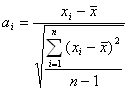
where xi is the original values for functional
group position index (P), carbon atom numbers (N), methyl numbers (Nm) and
boiling points, n is the number of the data, and ai was the result of the
standardization.
Table 2 Compounds selected in training set and testing set
Sets |
Compound codes as listed in Table 1 |
Training set |
56, 12, 35, 29, 44, 50, 33, 25, 64, 63, 20, 51, 66, 16, 46, 43, 13, 7, 59, 32, 45, 6, 71, 1, 60, 68, 2, 37, 17, 55, 19, 31, 48, 4, 52, 28, 54, 23, 38, 21, 18, 58, 73, 27, 47, 65, 41, 72, 57, 9. |
Testing set |
49, 10, 62, 22, 36, 34, 40, 26, 24, 42, 53, 67, 69, 15, 30, 61, 14, 8, 5, 70, 39, 3, 11 |
To finish the task of predicting boiling points of saturated aliphatic aldehydes and alkanones, the following procedures were done. First, we should choose the appropriate hidden layer nodes. Second, we compared the effect of different learning algorithm using in the network. Third, the prediction was done. Finally, we estimated the predicted results, and compared the BPNN method with other methods such as MLR method and topological index method previously reported.
3. RESULTS AND DISCUSSION
The neural network methodology has several empirically determined parameters. For
example, the number of hidden layer nodes, the number of training epochs or the
convergence criterion, the learning rate and momentum term, the initialization of the
network, and so on. After confirming the input and target of the network, network
optimization has to be done.
The training course will be ended when the mean square error (MSE)
values for training and testing sets simultaneously reaches the minimum. In the whole
training process, the MSE values for training and testing sets can be calculated, and
through monitoring the tendency of MSE we can determine if stopping the training process.
Generally, the MSE value for training set will constantly decrease with evolution of
training epochs. Similarly, the MSE value for testing set also decreases with evolution of
training epochs in the begining stage. But if continuing the training epochs, the MSE for
testing set will on the contrary increase, resulting in inferior prediction results for
testing set. To prevent the overtraining phenomenon, it is necessary to monitor
simultaneously the MSE for both training and testing sets.
Early stopping was used in the optimizing training process. In ANN,
early stopping [15, 16] is a pretty powerful and typical form using
cross-validation which is the widely used method to avoid the overtraining (or
over-fitting) phenomenon of neural networks. Early stopping means that the time stopping
training proceeds is controlled by the minimum of errors of the validation (or testing)
sets other than the minimum of errors of the training set. In general, the data set is
divided into training, validation and test sets, while in the case of a small data set,
the test set can substitute the validation set in cross-validation. So we divided the
studied data of 73 groups into two sets, training set and testing set.
3.1 BPNN model confirmation
There were 3 nodes in the input layer and
1 node in the output layer. Our first aim was to determine the optimal number of hidden
layer nodes. A
series of neural networks with different numbers of hidden layer nodes were trained. The number
of hidden layer nodes varied from 3 to 8. According to its generalization ability on the
testing set, we calculated the mean square error (MSE) on different numbers of the hidden
layer nodes. MSE is computed with the following equation:
![]()
where di is the desired output (the experimental
boiling point) in testing set, oi is the actual output in testing set, and n is
the number of compounds in the testing set. The lower the value of MSE, the better the
network model. To see the transformation trend intuitively, a curve of MSE versus the
number of hidden layer nodes was plotted (as shown in Fig. 3). Fig. 3 shows that the best
number of hidden layer nodes is 6. So a 3-6-1 BPNN model was selected for further studies.
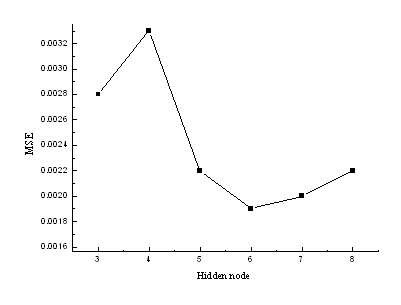
Fig. 3 Hidden node numbers vs mean
square error (MSE) on testing set of the saturated aliphatic aldehydes and alkanones
3.2 Learning algorithm comparison
Various learning rules derived from the first descent learning. Several different modifications of BP learning rule were selected in the training course. The learning epochs and the correlation coefficient of predicted boiling points and original experimental results are listed in Table 3. As shown in Table 3, L-M algorithm is the best learning rule for predicting boiling points of saturated aliphatic aldehydes and alkanones.
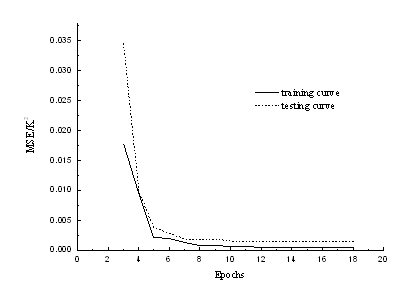
Fig. 4 The MSE of the training and testing set vs the learning epochs.
Table 3 Learning algorithm comparison
Learning algorithm* |
Training epochs |
Min MSE/K2 |
Correlation Coefficient (r) |
GDBP |
3617 |
0.00638 |
0.9969 |
GDABP |
177 |
0.0139 |
0.9941 |
GDMBP |
20000 |
0.00416 |
0.9973 |
GDXBP |
99 |
0.0196 |
0.9925 |
L-MBP |
14 |
0.000541 |
0.9993 |
*GDBP: gradient descent back-propagation; GDABP: gradient descent with adaptive learning rate (lr) back-propagation; GDMBP: gradient descent with momentum back-propagation; GDXBP: gradient descent with momentum & adaptive lr back-propagation; L-MBP: L-M back-propagation.
3.3 Predictions of boiling point using BPNN
As discussed above, we confirmed a 3-6-1 BPNN, and validated that the best learning
algorithm was L-M back-propagation. Fig. 5 is the plot of experimental boiling points
against predicted boiling points. Fig. 5 shows that almost each point falls on the
straight line of y=x, indicating that the predicted results are close to experimental
results. Linear regression showed that the predicted boiling points were in extremely good
agreement with those of the experimental data. The linear regression equation is given as:
![]()
where Tb,pre refers to the predicted boiling
point, and Tb,exp refers to the experimental boiling point. The correlation
coefficient (r) was 0.9992, indicating that the prediction was especially successful.
Twenty three boiling points in the testing set paired with the predicted results are given
in Table 4. The relative predicting error for boiling points is 1.8 %. The RMS error was
computed with the following equation:
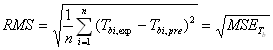
where Tbi,pre refers to the predicted result, and
Tbi,exp refers to the experimental boiling point, and n means the number of
compounds in the testing set. The RMS error of this prediction is 2.85 K, which is
significantly lower than the RMS error reported in the reference [4].
After predicting the testing set data, we simulated the training set
data using the optimized BPNN. Fig. 5 also illustrated that the predicted results were
perfect. Through linear regression, the linear regression equation was given as:
![]()
Its correlation coefficient is 0.9993,
indicating that the predicted results were also extremely close to the experimental
boiling points. The data are also listed in Table 4.
The residual errors between all the predicted and experimental boiling
points are showed in Fig. 6. As shown in Fig. 6 residual errors for boiling points
predicted by BPNN method were in the range from +9 K to -6 K. The boiling point of
formaldehyde was excluded in the previous reports due to its smaller molecular weight,
while in this report its absolute error is only 8.6 K and relative error is 3.2 %.
Although the absolute error for predicting boiling point of formaldehyde is the maximum in
our predicted results, this error falls in the acceptable range.
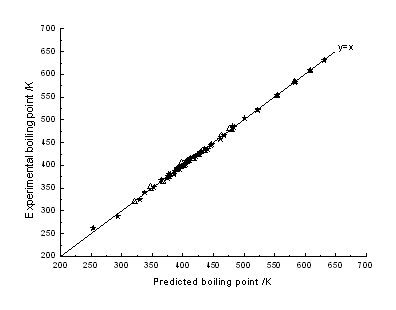
Fig.5 BPNN predicted vs experimental boiling
points of all the data.
3.4 Method comparison
For comparison with BPNN, MLR analysis was carried out using the number of carbon atom (N), the carbonyl position index (P) and the number of methyl (Nm) as variables. The obtained MLR equation was:
y=298.4188+19.0872×N - 16.7502×P - 4.7512×Nm
R2 was 0.9774, and RMS error was 11.6 K. The residual errors given by MLR were plotted in Fig. 6 (b). As shown in Fig. 6, the residual error by MLR is in the range from +50 K to -20 K, while the residual error by BPNN for most data points is in the range from +6 K to –6 K, only two exceptional cases for formaldehyde and 4,4-dimethyl-2-pentanone. These results were significantly worse than the results achieved in BPNN way.
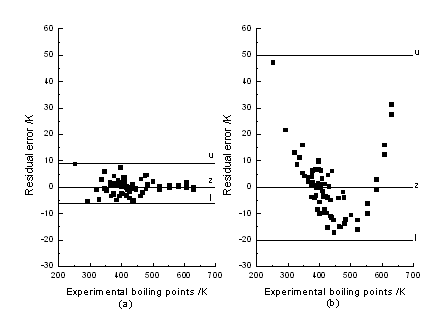
Fig. 6 Residual errors between all predicted and experimental boiling points. (a): BPNN method; (b): MLR method. u: upper limit of all data's residual error, l: lower limit of all data's residual error, z: line referred to residual error was zero.
Table 4 Experimental and predicted boiling points & errors of the methods BPNN and MLR
No. |
Tb,exp/K |
BPNN |
MLR |
||||
Tb,pre/K |
Error/K |
Relative Error/% |
Tb,pre/K |
Error/K |
Relative Error/% |
||
1 |
253.7 |
262.3 |
8.6 |
3.4 |
300.8 |
47.1 |
18.55 |
2 |
293.6 |
288.3 |
-5.3 |
-1.79 |
315.1 |
21.5 |
7.32 |
*3 |
321.1 |
320.2 |
-0.9 |
-0.27 |
334.2 |
13.1 |
4.07 |
4 |
329.4 |
324.6 |
-4.8 |
-1.45 |
337.8 |
8.4 |
2.55 |
*5 |
348 |
347.3 |
-0.7 |
-0.19 |
353.3 |
5.3 |
1.51 |
6 |
337.3 |
340.1 |
2.8 |
0.82 |
348.5 |
11.2 |
3.32 |
7 |
352.8 |
351.5 |
-1.3 |
-0.36 |
356.9 |
4.1 |
1.16 |
*8 |
376.2 |
373.7 |
-2.5 |
-0.67 |
372.4 |
-3.8 |
-1.02 |
9 |
365 |
366.7 |
1.7 |
0.46 |
367.6 |
2.6 |
0.71 |
*10 |
365.8 |
366.1 |
0.3 |
0.09 |
367.6 |
1.8 |
0.49 |
*11 |
347 |
352.9 |
5.9 |
1.67 |
362.9 |
15.9 |
4.57 |
12 |
375.5 |
377.0 |
1.5 |
0.4 |
376.0 |
0.5 |
0.13 |
13 |
375.1 |
372.1 |
-3.0 |
-0.8 |
378.8 |
3.7 |
0.99 |
*14 |
367.4 |
364.0 |
-3.4 |
-0.93 |
371.2 |
3.8 |
1.04 |
*15 |
401.5 |
399.1 |
-2.4 |
-0.61 |
391.4 |
-10.1 |
-2.51 |
16 |
390.2 |
391.8 |
1.6 |
0.41 |
386.7 |
-3.5 |
-0.90 |
17 |
395.2 |
391.8 |
-3.4 |
-0.86 |
386.7 |
-8.5 |
-2.15 |
18 |
395.2 |
391.8 |
-3.4 |
-0.86 |
386.7 |
-8.5 |
-2.15 |
19 |
377.2 |
381.4 |
4.2 |
1.1 |
381.9 |
4.7 |
1.26 |
20 |
386.2 |
381.4 |
-4.9 |
-1.26 |
381.9 |
-4.3 |
-1.10 |
21 |
380.2 |
381.4 |
1.2 |
0.3 |
381.9 |
1.7 |
0.46 |
*22 |
390 |
391.0 |
1.0 |
0.26 |
386.7 |
-3.3 |
-0.85 |
23 |
400.9 |
401.1 |
0.2 |
0.04 |
395.1 |
-5.8 |
-1.46 |
*24 |
396.7 |
398.0 |
1.3 |
0.34 |
397.9 |
1.2 |
0.31 |
25 |
390.6 |
393.2 |
2.6 |
0.67 |
390.3 |
-0.3 |
-0.07 |
*26 |
389.6 |
390.0 |
0.4 |
0.09 |
390.3 |
0.7 |
0.18 |
27 |
386.6 |
387.4 |
0.8 |
0.2 |
393.2 |
6.6 |
1.70 |
28 |
379.5 |
380.2 |
0.7 |
0.19 |
385.6 |
6.1 |
1.60 |
29 |
426 |
424.1 |
-1.9 |
-0.44 |
410.5 |
-15.5 |
-3.63 |
*30 |
415.7 |
415.3 |
-0.4 |
-0.09 |
405.8 |
-9.9 |
-2.39 |
31 |
424.1 |
423.7 |
-0.4 |
-0.09 |
414.2 |
-9.9 |
-2.35 |
32 |
420.6 |
419.7 |
-0.9 |
-0.21 |
417.0 |
-3.6 |
-0.86 |
33 |
417.2 |
417.3 |
0.1 |
0.01 |
418.3 |
1.1 |
0.27 |
*34 |
413 |
414.7 |
1.7 |
0.82 |
409.4 |
-3.6 |
-0.87 |
35 |
412 |
414.7 |
2.7 |
0.66 |
409.4 |
-2.6 |
-0.63 |
*36 |
418 |
414.7 |
-3.30 |
-0.8 |
409.4 |
-8.6 |
-2.06 |
37 |
406 |
409.5 |
3.50 |
0.85 |
412.2 |
6.2 |
1.54 |
38 |
409 |
409.5 |
0.50 |
0.11 |
412.2 |
3.2 |
0.79 |
*39 |
409 |
409.1 |
0.1 |
0.03 |
412.2 |
3.2 |
0.79 |
*40 |
403.8 |
405.4 |
1.60 |
0.38 |
404.6 |
0.8 |
0.21 |
41 |
405 |
402.4 |
-2.70 |
-0.65 |
404.6 |
-0.4 |
-0.09 |
*42 |
398 |
405.4 |
7.40 |
1.81 |
404.6 |
6.6 |
1.67 |
43 |
411 |
414.7 |
3.70 |
0.91 |
409.4 |
-1.6 |
-0.39 |
44 |
398 |
398.4 |
0.40 |
0.1 |
407.5 |
9.5 |
2.39 |
45 |
397.6 |
398.4 |
0.80 |
0.2 |
407.5 |
9.9 |
2.49 |
46 |
447 |
446.0 |
-1.00 |
-0.23 |
429.6 |
-17.4 |
-3.89 |
47 |
436 |
436.9 |
0.90 |
0.22 |
424.9 |
-11.1 |
-2.55 |
48 |
445.8 |
445.1 |
-0.70 |
-0.16 |
433.2 |
-12.6 |
-2.82 |
*49 |
463.7 |
466.6 |
2.90 |
0.61 |
448.7 |
-15.0 |
-3.23 |
50 |
440.2 |
435.1 |
-5.10 |
-1.15 |
428.5 |
-11.7 |
-2.66 |
51 |
440.2 |
435.1 |
-5.10 |
-1.15 |
428.5 |
-11.7 |
-2.66 |
52 |
431.2 |
430.6 |
-0.60 |
-0.14 |
431.3 |
0.1 |
0.03 |
*53 |
436.2 |
431.1 |
-5.10 |
-1.19 |
431.3 |
-4.9 |
-1.12 |
54 |
428.2 |
428.1 |
-0.1 |
-0.02 |
432.7 |
4.5 |
1.05 |
55 |
427.7 |
423.4 |
-4.3 |
-1.01 |
423.7 |
-4.0 |
-0.93 |
56 |
467.5 |
465.3 |
-2.20 |
-0.47 |
452.3 |
-15.2 |
-3.25 |
57 |
461.6 |
458.2 |
-3.40 |
-0.73 |
457.4 |
-4.2 |
-0.92 |
58 |
441 |
436.2 |
-4.9 |
-1.1 |
447.0 |
6.0 |
1.36 |
59 |
481.7 |
486.1 |
4.40 |
0.92 |
467.8 |
-13.9 |
-2.89 |
60 |
483.7 |
484.6 |
0.90 |
0.19 |
471.4 |
-12.3 |
-2.54 |
*61 |
476.2 |
480.5 |
4.30 |
0.89 |
474.3 |
-1.9 |
-0.41 |
*62 |
479.7 |
478.9 |
-0.8 |
-0.17 |
475.6 |
-4.1 |
-0.85 |
63 |
501.2 |
503.2 |
2.00 |
0.39 |
490.5 |
-10.7 |
-2.13 |
64 |
522.2 |
522.2 |
0.00 |
-0.01 |
506.0 |
-16.2 |
-3.11 |
65 |
522.2 |
521.1 |
-1.1 |
-0.2 |
509.6 |
-12.6 |
-2.42 |
66 |
554.2 |
554.6 |
0.40 |
0.07 |
544.1 |
-10.1 |
-1.82 |
*67 |
554.2 |
553.7 |
-0.50 |
0.1 |
547.8 |
-6.4 |
-1.16 |
68 |
583.2 |
583.6 |
0.40 |
0.06 |
582.3 |
-0.9 |
-0.15 |
*69 |
583.2 |
583.1 |
-0.10 |
0.02 |
585.9 |
2.7 |
0.47 |
*70 |
608.2 |
608.0 |
-0.20 |
-0.03 |
620.5 |
12.3 |
2.02 |
71 |
608.2 |
609.9 |
1.70 |
0.28 |
624.1 |
15.9 |
2.62 |
72 |
631.2 |
631.1 |
-0.10 |
-0.02 |
658.7 |
27.5 |
4.35 |
73 |
631.2 |
630.3 |
-0.90 |
-0.15 |
662.3 |
31.1 |
4.92 |
BPNN |
*RMS/K |
2.85 |
|||||
MLR |
RMS/K |
11.59 |
|||||
*belonging to testing set.
A comparison of our work with topological index methods previously reported was also made in terms of descriptor number, correlation coefficient (r) and the number of data set. The comparison results were given in Table 5. The BPNN method proposed in this work only needs three simple descriptors, which have exact chemical meaning, while topological methods [4-6, 8] previously reported need many descriptors obtained by complicated calculation procedures.
Table 5 Comparison of different boiling points predicting methods
Method |
Number and type of descriptor |
Correlation coefficient |
Reference |
BPNN |
3: carbon atom numbers; carbonyl position index; methyl numbers |
r=0.9992 |
Our work |
Topological index |
12: extended molecular distance-edge (MDE, μ) vector, m1; m2; m3; m4; m5; m6; m7; m8; m9; m10; m12; m14 |
R=0.9989 |
[4] |
Topological index |
3: topological index of atomic ordinal number mM, carbon atom numbers; 0M; 1M |
R=0.9991 |
[5] |
Topological index |
3: effective length of carbon chain; carbon atom numbers; inductive effect index difference between the corresponding branched and normal alkyl isomer containing the same carbon atom number |
R=0.9987 |
[6] |
Topological index |
3: connectivity index 1Q; converse index 1Q'; the largest point valence of carbon atom dmax |
R=0.9990 |
[8] |
3.5 Network structure validation
The 3-6-1 BPNN structure was built to predict the boiling points of 73 saturated
aliphatic aldehydes and alkanones. The stability of the network structure was validated
through several different random data grouping. Except the training set and testing set
used above, the other 4 random divided situations were generated for predicting boiling
points. Linear regression was done for the original boiling points and the predicted
results obtained from these parallel experiments. The specific components in training and
testing sets for 5 times of parallel experiments and their correlation coefficients for
the original boiling points and the predicted results obtained from these parallel
experiments were given in Table 6. The average of correlation coefficients for the
original boiling points and the predicted results obtained from these parallel experiments
was 0.9992, indicating that the 3-6-1 BPNN is stable and suitable for the prediction of
boiling points of the studied 73 saturated aliphatic aldehydes and alkanones.
Table 6 Model validation
Divided situation |
Compound numbers |
r |
1 |
Shown in Table 2 |
0.9992 |
2 |
Training set: 26, 69, 40, 61, 59, 52, 18, 65, 63, 46, 11, 37, 60, 50, 30, 35, 44, 2, 1, 43, 72, 21, 55, 39, 20, 58, 56, 67, 12, 19, 17, 36, 8, 51, 16, 6, 13, 32, 64, 25, 54, 62, 15, 27, 70, 48, 31, 3, 34, 14 |
0.9996 |
Testing set: 41, 23, 66, 4, 9, 28, 68, 10, 49, 53, 22, 57, 73, 42, 7, 45, 33, 47, 71, 24, 38, 29, 5 |
||
3 |
Training set: 19, 51, 47, 45, 26, 72, 18, 12, 21, 73, 38, 69, 58, 33, 15, 65, 14, 24, 22, 49, 60, 31, 2, 39, 63, 48, 29, 32, 56, 66, 5, 42, 25, 57, 30, 10, 36, 4, 43, 1, 34, 28, 46, 7, 71, 37, 59, 23, 41, 68 |
0.9985 |
Testing set: 6, 3, 62, 17, 64, 35, 52, 40, 55, 44, 8, 70, 50, 16, 11, 20, 67, 9, 61, 53, 13, 27, 54 |
||
4 |
Training set: 38, 7, 45, 46, 36, 42, 35, 3, 49, 1, 54, 41, 64, 28, 58, 47, 29, 25, 14, 71, 69, 21, 68, 5, 37, 16, 57, 50, 32, 39, 19, 4, 12, 22, 56, 65, 34, 55, 62, 13, 63, 33, 48, 2, 30, 44, 43, 27, 31, 51 |
0.9991 |
Testing set: 15, 59, 40, 24, 53, 60, 66, 52, 18, 61, 20, 8, 11, 72, 26, 70, 6, 67, 73, 23, 17, 9, 10 |
||
5 |
Training set: 5, 73, 12, 23, 2, 72, 6, 36, 66, 31, 32, 8, 11, 20, 7, 69, 68, 10, 49, 34, 30, 53, 50, 48, 3, 56, 38, 25, 17, 29, 59, 14, 16, 42, 27, 64, 19, 51, 35, 58, 55, 41, 9, 61, 45, 47, 21, 24, 33, 60 |
0.9994 |
Testing set: 37, 57, 28, 52, 71, 13, 62, 4, 46, 44, 26, 22, 54, 18, 40, 39, 70, 65, 1, 43, 15, 63, 67 |
||
Average r |
0.9992 |
|
4.CONCLUSION
The results obtained in this paper demonstrate that it is possible to generate robust
networks capable of estimating the boiling points of saturated aliphatic aldehydes and
alkanones using functional group position index (P), carbon atom numbers (N) and methyl
numbers (Nm) as inputs. The advantage of this work performed here as compared
with other methods is that no experimental parameters are required and the selected three
parameters are easily obtained from the molecular structures for saturated aliphatic
aldehydes and alkanones. The BPNN proposed in this work has been shown to provide more
accurate prediction of boiling points than those through linear regression analysis
approach.
ACKNOWLEDGEMENTS Partially financial support from the National Natural Science Foundation of China (20575016) and the Natural Science Foundation of Hebei Province China (B2006000953) are gratefully acknowledged.
REFERENCES
[1]Balaban A T, Mills D, Basak S C. Journal of Chemical Information and Computer Sciences,
1999 39 (4): 758-764.
[2]Toropov A A, Toropova A P. Jounal of Molecular Structure: Theochem, 2002, 581 (5):
11-15.
[3]Toropov A A, Toropova A P, Mukhamedzhanova D V et al. Indian Journal of Chemistry –
Section A Inorganic, Physical, Theoretical and Analytical
Chemistry, 2005, 44 (8): 1545-1552.
[4]Lin Zhihua, Xu Jianghe, Liu Shushen et al. Acta Physico-Chemica Sinica, 2000, 16 (2):
153-161.
[5]Chen Yan. Chinese Journal of Organic Chemistry, 2001, 21 (3): 242-246.
[6]Zhang Xiuli, Wang Yongxian, Li Junling et al. Chinese Journal of Organic Chemistry,
2002, 22 (11): 897-901.
[7]Zhang Xiuli, Wang Yongxian, Li Junling et al. Chemical Research and Application, 2003,
15 (6): 803-804.
[8]Feng Changjun, Yang Weihua. Journal of Jilin University (Science Edition), 2003, 41
(1): 97-101.
[9]Wang Keqiang. Chinese Journal of Organic Chemistry, 1998, 18: 419-424.
[10]Chen Gang, Hu Fang, Xiang Jiannan et al. Journal of Hunan University, 1998, 25 (1):
27-30.
[11]Zhang Xiangdong, Zhao Lijun, Zhang Guoyi. Chemical Research and Application, 1994, 6
(3): 48-53.
[12]Yannis L. Loukas. Journal of Chromatography A, 2000, 904: 119-129.
[13]Jalali-Heravi M., Garkani-Nejad Z.. Journal of Chromatography A, 2002, 945: 173-184.
[14]Ma Shichang. Chemical Substances Dictionary. 1. Shanxi: science and technology
publishing company, 1994.
[15]Finnoff W, Hergert F, Zimmermann H G. Neural Networks, 1993, 6: 711.
[16]Lang K J, Waibel A H, Hinton G E. Neural Networks, 1990, 3: 33.
成谦,张红医
(化学与环境科学学院,河北大学,保定071002,中国)
摘要 利用3-6-1型反向传播的人工神经网络(BPNN)对73个饱和脂肪醛酮及沸点进行了定量结构-性质关系研究。根据饱和脂肪醛酮的结构,结合系统命名法建立了官能团位置参数P,建立3-6-1型BP网络,采取P、碳原子数(N)和甲基个数(Nm)作为网络的输入。以此方法预测沸点,并得到了良好的结果。将预测结果与文献值作回归分析,相关系数r为0.9992,RMS为2.9 K。本文采用的预测方法简便易行,仅需要3个简单的结构描述码即可得到良好的预测值,适宜用来预测饱和脂肪醛酮的沸点。
关键词:定量结构-性质关系,人工神经网络,沸点, 饱和脂肪醛, 饱和脂肪酮