http://www.chemistrymag.org//cji/2008/102008ne.htm |
Feb. 1,
2008 Vol.10 No.2 P.8 Copyright |
Synthesis, characterization and crystal structure of a new binuclear copper(II) complex with indole-3-propionic acid
Deng Xiujun, Bian Hedong, Yu Qing,
Liang Hong
(College of Chemistry and Chemical Engineering, Guangxi Normal University, Guilin 541004,
China)
Supported by Teaching and Research Program for Outstanding Young Teachers in Higher Education Institutions of Ministry of Education, the Nature Science Foundation of Guangxi Province and the Nature Science Foundation of Guangxi Normal University.
Abstract A binuclear copper(II) complex, Cu2(IPA)4(CH3OH)2 (1) (HIPA = indole-3-propionic acid), was synthesized and characterized by IR, elemental analysis and X-ray diffraction. The crystal of 1 belongs to Triclinic system, space group PKeywords indole-3-propionic acid, crystal structures, binuclear copper complex
1. INTRODUCTION
Indole-3-propionic acid (HIPA), a deamination product of tryptophan is formed by symbiotic
bacteria in the gastrointestinal tract of mammals and birds[1]. HIPA may hold
particular promise as a therapeutic agent in human brain diseases[2]. It was
found that anti-inflammatory and antibacterial activity of metal complexes was higher than
in the parent carboxylic acids[3]. The carboxylate group displays a variety of
binding geometries, such as monodentate, chelating, bidentate bridging and monodentate
bridging so in coordination chemistry as well as in the active sites of metalloenzymes[4-8]. It should be noted that the crystal structures of HIPA
have been studied[9,10], but there are seldom reports about the crystal
structure of its metal complexes[11], as yet. In the present paper, Cu2(IPA)4(CH3OH)2
was synthesized and characterized by IR, elemental analysis, and X-ray diffraction.
2. EXPERIMENTAL
2.1 Instruments and chemicals
IR spectrum was recorded on a Perkin-Elmer Spectrum one FT-IR Spectrometer in the 4000
-400 cm-1 region by using KBr pellets. Elemental analysis for C, H and N atoms
were carried out on a Model 2400 II Perkin-Elmer elemental analyzer. All reagents were of
analytical grade from commercial sources and were used without any further purification.
2.2 Synthesis of Cu2(IPA)4(CH3OH)2 (1)
HIPA was dissolved in 5 mL of methanol and the pH value was adjust to about 7 with 1
mol·L-1 NaOH solution. Then the solution of Cu(NO3)2·3H2O
(0.5 mmol) in 10 mL of methanol was added. The mixture was stirred at 60
IR (KBr) n:. 3411.64(vs), 1606.18(s), 1423.69(vs), 1384.79(ms), 1358.64(w), 1226.90(w), 1094.85(m), 1010.66(m), 740.23(s), 613.77(m), 493.85(m)cm-1.
Anal. calcd for C46H48Cu2N4O10: C, 58.48; H, 5.13; N, 5.93%. found: C, 57.62; H, 5.52; N, 5.86%.
2.3 X-ray crystallography of 1
Table 1 Crystal data and structure refinement for complex 1
Empirical formula |
C46H48Cu2N4O10 |
Absorption coefficient(mm-1) |
1.020 |
Formula weight |
943.96 |
F(000) |
980 |
Crystal system |
Triclinic |
Crystal size (mm3) |
0.21 × 0.17 × 0.07 |
space group |
P |
q (°) | 1.41 to 25.01 |
a (nm) |
0.7521(2) |
Limiting indices |
-8≤h≤8, -18≤k≤13, -24≤l≤21 |
b (nm) |
1.5799 (3) |
Reflections collected |
10895 |
c (nm) |
2.0432(3) |
Independent reflections | 7338 |
| a (°) | 68.330(2) | Data / restraints / parameters |
7338 / 18/ 559 |
| b (°) | 85.804(3) | Goodness-of-fit on F2 |
0.993 |
| g (°) | 79.622(2) |
R [I>2s(I)] |
0.0995 |
Volume (nm3 ) |
2.2192(8) |
wR2 |
0.1555 |
Z |
2 |
Maximum diff. peak (e/nm3) |
1.248 |
Dc (g/cm3) |
1.413 |
Minimum diff. peak (e/nm3) |
-1.351 |
3 RESULTS AND DISCUSSION
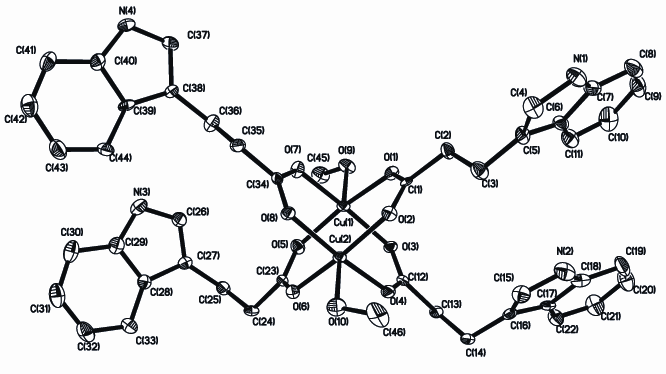
Fig. 1 The
molecular structure of Cu2(IPA)4(CH3OH)2 (H
atoms are omitted for clarity)
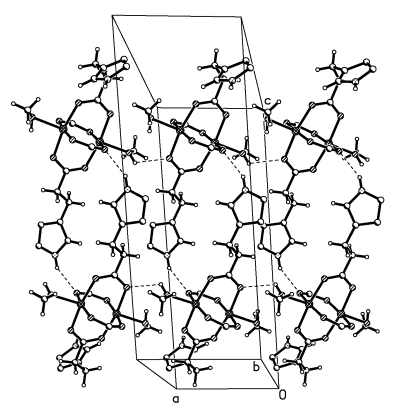
Fig. 2 the 1D band diagram of the title compound along a-axis
(Some atoms have been omitted for carily)
Table 2 Selected bond lengths (nm) and bond angles (o)
| Bond length | |||
| Cu(1)-O(5) | 0.1963(7) | Cu(2)-O(6) | 0.1960(7) |
| Cu(1)-O(1) | 0.1967(8) | Cu(2)-O(4) | 0.1976(7) |
| Cu(1)-O(7) | 0.1974(7) | Cu(2)-O(2) | 0.1987(7) |
| Cu(1)-O(3) | 0.1991(7) | Cu(2)-O(8) | 0.2001(7) |
| Cu(1)-O(9) | 0.2156(7) | Cu(2)-O(10) | 0.2191(7) |
| Cu(1)-Cu(2) | 0.26221(17) | ||
| Bond angle | |||
| O(5)-Cu(1)-O(1) | 167.5(3) | O(6)-Cu(2)-O(4) | 89.6(3) |
| O(5)-Cu(1)-O(7) | 91.0(3) | O(6)-Cu(2)-O(2) | 170.0(3) |
| O(1)-Cu(1)-O(7) | 89.0(3) | O(4)-Cu(2)-O(2) | 89.4(3) |
| O(5)-Cu(1)-O(3) | 90.9(3) | O(6)-Cu(2)-O(8) | 88.5(3) |
| O(1)-Cu(1)-O(3) | 86.7(3) | O(4)-Cu(2)-O(8) | 168.4(3) |
| O(7)-Cu(1)-O(3) | 168.6(3) | O(2)-Cu(2)-O(8) | 90.6(3) |
| O(5)-Cu(1)-O(9) | 97.7(3) | O(6)-Cu(2)-O(10) | 93.2(3) |
| O(1)-Cu(1)-O(9) | 94.7(3) | O(4)-Cu(2)-O(10) | 95.9(3) |
| O(7)-Cu(1)-O(9) | 97.0(3) | O(2)-Cu(2)-O(10) | 96.8(3) |
| O(3)-Cu(1)-O(9) | 93.8(3) | O(8)-Cu(2)-O(10) | 95.6(3) |
Table 3 Hydrogen-bonding geometry (nm, °) for the title complex
D-H |
d(D-H) |
d(H..A) |
<DHA |
d(D..A) |
A |
N4-H4 |
0.860 |
2.151 |
151.28 |
2.934 |
O1[-x+3, -y+3, -z+1] |
O9-H9 |
0.820 |
2.452 |
118.53 |
2.934 |
O8[ x+1, y, z ] |
O10-H10 |
0.820 |
2.484 |
133.86 |
3.109 |
O3[ x, y, z ] |
REFERENCES
[1] Young S, Anderson G M, Gauthier S et al. J. Neurochem, 1980, 34: 1087.
[2] Poeggeler B, Pappolla M A, Hardeland R et al. Brain Res, 1999, 815: 382.
[3] Dendrinou-Samara C, Tsotsou G, Ekateriniadou L V et al. J. Inorg. Biochem, 1998, 71:
171.
[4] Ye B H, Chen X M., Xue F et al. Inorg. Chim. Acta, 2000, 299: 1.
[5] Lemoine P, Viossat B, Huy D N. J.Inorg. Biochem, 2004, 98: 1734.
[6] Bresolin L, Burow R A, H?rner M et al. Polyhedron, 1997, 16: 3947.
[7] Cui Y, Qian Y T, Huang J S, Polyhedron, 2001, 20: 1795.
[8] Yin M C, Yuan L J, Ai C C et al. Polyhedron, 2004, 23: 529.
[9] Koshima H, Hayashi E, Matsuura T et al. Tetrahedron Lett, 1997, 38: 5009.
[10] Okabe N, Adachi Y, Acta Cryst. C, 1998, 54: 386.
[11] Barbara M O, Ewa R S. Vibrational Spectroscopy, 2007, 43: 405.
[12] Sheldrick G M SHELXS-97, Program for X-ray Crystal Structure Solution [M]. University
of Göttingen : Germany, 1997.
[13] Sheldrick G M SHELXL-97, Program for the Refinement of Crystal Structures [M].
University of G
[14] Wu B, Wang G C. Acta Cryst. E, 2004, 60: m1764
[15] Wang Y Y, Shi Q, Shi Q Z et al. Polyhedron, 2000, 19: 891.
[16] Benisvy L, Quesada M, Turpeinen U et al. Chem. Commun, 2006: 2373.
[17] Calvo P V, Vega A, Spodine E. Organometallics, 2006, 25: 1953
[18] Deka K, Barooah N, Sarma R J et al. J. Mol. Struct, 2007: 44.
[19] Addison A W, Rao T N, Reedijk J et al. J. Chem. Soc. Dalton Trans., 1984: 1349. 一个新型的吲哚-3-丙酸双核铜配合物的合成和晶体结构表征
邓秀君 边贺东 于青 梁宏
(广西师范大学化学化工学院,桂林 541004,中国)
摘要 本文合成了一个新型的双核配合物Cu2(IPA)4(CH3OH)2(HIPA=吲哚-3-丙酸),并用红外、元素分析、X射线衍射等方法对其结构进行表征。该单晶属三斜晶系,空间群为P
关键词 吲哚-3-丙酸,晶体结构,双核铜配合物