http://www.chemistrymag.org/cji/2002/048038pe.htm |
May 20,
2002 Vol.4 No.8 P.38 Copyright |
A fast algorithm based on wavelet compression and immune algorithm for resolution and quantitative determination of the component in multicomponent overlapping chromatogram
ShaoXueguang,
Yu Zhengliang
(Department of Chemistry, University of Science and Technology of China, Hefei, Anhui,
230026, China)
Received Feb.20, 2002; Supported by the National Natural Science Foundation of China (No.29975027)
Abstract Based on the wavelet compression
and immune algorithm (IA), a novel algorithm for fast resolution of two-dimensional
multicomponent overlapping chromatogram is proposed. Due to the characteristic of the
linear property of the wavelet transform (WT), the overlapping chromatogram (antigen) can
be compressed by WT before it is input into the immune network, the standard chromatogram
of each component (antibodies) is also compressed to the same scheme. Especially, for
speeding up the computation, the two-dimensional data matrix is arranged into
one-dimensional vector form before it is compressed. After the compressed information of
each component was extracted by IA, the chromatogram can be reconstructed by the inverse
WT algorithm and re-arranged back into matrix form. It was proven that the result is
almost the same with the result from IA, but the calculation speed is much faster. At the
same time, satisfactory quantitative result can also be obtained.
Keywords Immune algorithm, Wavelet transform, Compression, Resolution.
1. INTRODUCTION
Along with the development of modern chemical instrumentation, multicomponent
two-dimensional data matrices can be easily obtained. For the aim of resolving the
multicomponent overlapping matrices, several methods, such as chemical factor analysis
(CFA),[1,2] wavelet transform (WT) [3-5] and immune algorithm (IA) [6-8]
have been proposed. In our previous works,[6-10] it has been proven that the IA
is an efficient tool for the resolution of overlapping multicomponent analytical signals.
Multicomponent overlapping chromatogram can be easily resolved by an IA and the
calculation speed is faster than conventional least-square method.[7] However,
when the size of data set is large and there are parameters need to be optimized, the
consumed time of the computation is too long to be feasible in practical uses, e.g., in
GA-IA[10] method, because the IA procedure is invoked repeatedly. One useful
way to speed up the calculation is to compress the raw experimental data. There have been
many efficient tools for analytical data compression, such as binary coding method, Adams
and Black algorithm, Fourier transform, chemical factor analysis, and wavelet transform.
[11-14]
In this paper, both the matrices of the standard
chromatogram (antibodies) and the multicomponent overlapping chromatogram (antigen) are
converted into one-dimensional vector form and compressed by wavelet transform at first,
then perform the resolution by using an IA. It was found that the calculation speed can be
improved by the conversion and compression. It can provide a fast preprocessing tool to
the GA-IA method.
2. ALGORITHM
The principle and application of IA has been reported in our previous works.[6-10]
The essence of an IA is that, taking the signal of the multicomponent mixture as antigen
and the signals of the standard samples as antibodies, the information of each component
in multicomponent overlapping signal can be extracted by a process of recognition,
iterative elimination etc. The calculation process of an IA can be simply described by the
following formulae:
![]() (1)
(1)
![]() (2)
(2)
![]() (3)
(3)
where T is the weight of input layer, k is the number of iteration, V
is the overlapping chromatographic signal (antigen), V0i is the
standard chromatographic signal of the ith component with known concentration
(antibodies), ci is the relative concentration of the ith
component, and VF is the feedback vector or matrix denoting the
eliminated antigen. It can be seen that when dc(k) approaches to zero, VF
will be the information of each component in the overlapping chromatographic signal. In
many cases, due to the variation caused by the experimental reproducibility etc.,
parameters of V0i, such as the position and the shape of the peaks may
need to be optimized. The optimization will be a time-consumed procedure when the data
number of V and V0i is large. Therefore, an efficient way to
compress the V and theV0i will be necessary for speeding up the
algorithm.
The wavelet transform has been proven to be an efficient technique for
analytical data compression.[15,16] The dual localization characteristic in
both frequency and time domains, the linearity, and the existence of fast algorithm make
the WT an ideal candidate for preprocessing data for the IA. In this paper, the
multi-resolution signal decomposition (MRSD) algorithm[17,18] is used.
Based on the algorithm of the IA and WT compression, a fast algorithm
for resolution of multicomponent overlapping chromatogram is proposed. The flowchart can
be described in Figure 1, including the following steps:
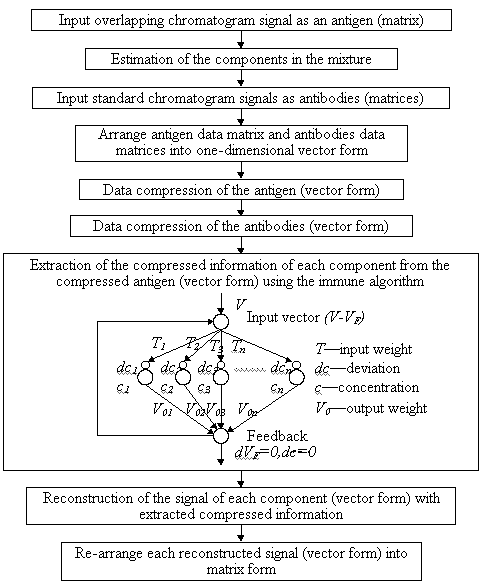
Fig.1 The flowchart of the proposed algorithm
(1) Input the overlapping 2-D chromatographic data matrix as antigen.
(2) Estimate the possible components from the original chromatogram.
(3) Input the standard 2-D chromatographic data matrices as antibodies.
(4) Arrange all 2-D data matrices of the antigen and antibodies into one-dimensional
vectors along with the chromatographic orientation.
(5) Apply the wavelet compression to the antigen vector, i.e., perform WT on the antigen
vector, then suppress those coefficients whose value is less than a threshold. The value
of the threshold is determined by a predefined compression ratio that is determined by
trial and inspection of the reconstruction error. The remained coefficients will be taken
as the antigen for further calculation.
(6) Compress the antibody vectors by performing WT with the same parameters as in the last
step and remaining those coefficients at the same position with the compressed antigen.
(7) Extract the compressed information of each component from the compressed antigen by
the IA mentioned above, where the compressed antigen is taken as V and the
compressed antibodies are taken as V0i. In this step, the compressed
information of each component can be extracted from the compressed antigen.
(8) Reconstruct the extracted information of each component by the inverse WT algorithm to
obtain a full extracted chromatographic data in vector form.
(9) Finally, re-arrange the extracted data vector back into matrix form.
After all above calculation, the chromatograms of each component can be resolved from the multicomponent overlapping data matrix. From the theory of the IA, ci is the concentration of each component relative to the concentration of the standard sample, i.e., if the concentration of the standard sample is known, then the concentration of each component in the mixture can be calculated by the parameter ci. Therefore, this method can give us both the resolution and the quantitative determination simultaneously.
3. EXPERIMENTAL
The experimental data sets were measured on an HPLC system comprising a Spectrasystem
FL2000 (Spectra-Physics, USA) with the spectra Focus multi-wavelength UV detector
(Spectra-Physics) and a Spectrasystem workstation. The column was packed with 10mm ODS silica (250 mm×5 mm, Shimadzu).
The mobile phase was 0.25mol/L (pH~3.5) lactic acid (A.R.) with 0.01mol/L dodecyl sulfonic
acid sodium. The color developing reagent of post column was 1.0×10-4mol/L
arsenazoⅢ (Fluka Chemie AG). The flow rate of the
mobile phase was 1.0mL/min. The flow rate of the color developing reagent was 1.0mL/min.
The temperature of the column was 20°C. The detection wavelength was from 580nm to 720nm.
The interval of sampling time is 0.005min.
Table 1 Composition of the samples (unit: mg/ml)
Sample No. |
Er |
Tm |
Yb |
1 |
0.2000 |
0.1999 |
0.2001 |
2 |
0 |
0.1499 |
0.2001 |
3 |
0 |
0.1999 |
0.2001 |
Table 1 shows the
composition of the three samples, which are mixed by Yb, Tm and Er, respectively. Figure 2
and Figure 3 (a) (b) (c) show the two-dimensional chromatogram of the mixture sample 1
(antigen) and the standard chromatograms of Yb, Tm and Er (antibodies) obtained by the
experiment.
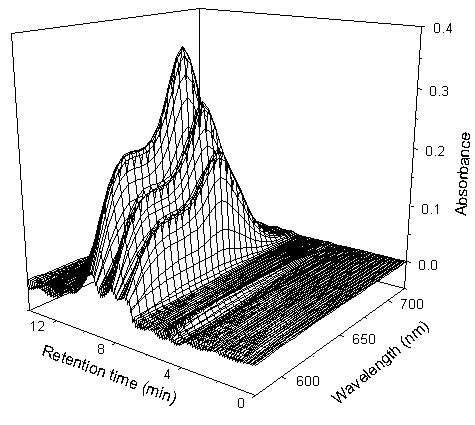
Fig.2 The experimental multicomponent overlapping chromatogram (antigen) of the sample
No.1
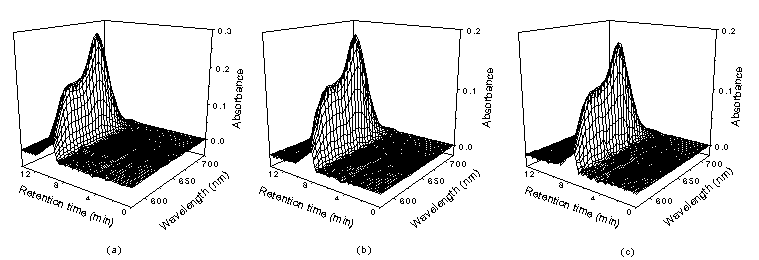
Fig.3 The standard chromatograms of single component (antibodies)
(a) Er (b) Tm (c) Yb
4. RESULTS AND DISCUSSION
4.1 Selection of the wavelet basis and the decomposition level
Figure 4 shows the coefficients obtained by the wavelet transform of the chromatogram
(arrange in vector form) in Figure 2 at level 7 using the Symmlet5 (L=10) wavelet
basis. It can be found that the information of the chromatographic signal is mainly
concentrated on only a few of the coefficients. Removing the smaller coefficient will not
affect the total information. In order to obtain the optimal wavelet basis and
decomposition level, reconstruction error obtained by Haar, Daubechies (L=4-20),
Coiflets (L=4-20), Symmlets (L=6-30) at different decomposition level, where
L is the length of filter, was investigated with the data sets in Figures 2 and 3. Table 2
summarized some of the results when the compression ratio is 1/58. The reconstruction
error is calculated by
![]() (4)
(4)
where, X is the original signal, XR is the reconstructed
signal from remained coefficients, n is the size of the original data set.
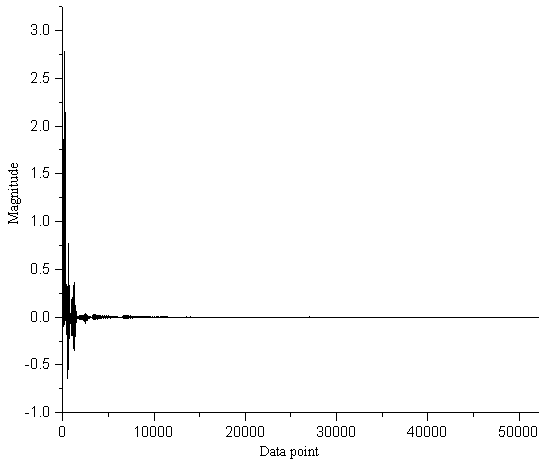
Fig.4 The wavelet coefficients obtained by WT of the chromatogram in Figure 2
From Table 2, it can be found that the variation of reconstructed error is almost the same for all the four data sets. For every wavelet basis the minimal reconstruction errors generally appear at decomposition level 6 or 7. Comparing the reconstructed errors between different wavelet basis, it can be found that Symmlet 4, 5, 6 give smaller results. Therefore, Symmlet5 at decomposition level 7 is adopted in the following studies.
Table 2 Reconstruction errors by different wavelet basis at different decomposition level
Wavelet basis |
Data |
Decomposition level |
||||
4 |
5 |
6 |
7 |
8 |
||
Haar |
Mix. |
309.601 |
194.953 |
177.447 |
171.839 |
168.520 |
Er |
94.252 |
78.993 |
74.375 |
73.626 |
74.169 |
|
Tm |
69.741 |
58.817 |
55.637 |
55.588 |
56.266 |
|
Yb |
69.070 |
58.046 |
53.986 |
52.874 |
51.045 |
|
Db4 |
Mix. |
276.538 |
114.605 |
92.823 |
88.633 |
95.276 |
Er |
68.043 |
43.651 |
37.795 |
37.023 |
40.095 |
|
Tm |
51.618 |
34.362 |
31.237 |
32.520 |
33.644 |
|
Yb |
53.556 |
37.539 |
32.522 |
30.989 |
32.992 |
|
Db8 |
Mix. |
277.929 |
109.555 |
89.389 |
88.817 |
99.135 |
Er |
68.060 |
42.529 |
38.665 |
40.783 |
46.580 |
|
Tm |
51.735 |
34.138 |
32.182 |
35.101 |
38.922 |
|
Yb |
53.529 |
37.544 |
33.170 |
33.769 |
37.752 |
|
Sym3 |
Mix. |
276.373 |
120.943 |
98.082 |
92.855 |
94.883 |
Er |
68.186 |
44.532 |
37.727 |
38.435 |
39.737 |
|
Tm |
51.816 |
35.000 |
31.922 |
32.380 |
34.307 |
|
Yb |
53.856 |
38.130 |
33.074 |
31.258 |
32.033 |
|
Sym4 |
Mix. |
275.467 |
115.130 |
92.764 |
85.817 |
87.348 |
Er |
67.950 |
43.506 |
37.304 |
37.294 |
40.324 |
|
Tm |
51.568 |
34.594 |
30.649 |
31.270 |
32.425 |
|
Yb |
53.539 |
37.624 |
32.082 |
30.208 |
30.807 |
|
Sym5 |
Mix. |
279.138 |
112.817 |
89.699 |
84.277 |
84.930 |
Er |
68.870 |
44.143 |
38.136 |
38.141 |
39.722 |
|
Tm |
52.365 |
35.039 |
31.369 |
31.726 |
31.908 |
|
Yb |
54.374 |
38.296 |
32.159 |
29.435 |
29.962 |
|
Sym6 |
Mix. |
275.515 |
110.207 |
87.794 |
82.778 |
86.383 |
Er |
67.938 |
42.396 |
36.546 |
37.034 |
39.834 |
|
Tm |
51.502 |
33.856 |
30.895 |
31.911 |
33.564 |
|
Yb |
53.468 |
37.125 |
32.402 |
30.549 |
31.978 |
|
Sym7 |
Mix. |
279.568 |
110.628 |
87.745 |
84.261 |
87.822 |
Er |
68.977 |
43.888 |
37.681 |
38.089 |
40.615 |
|
Tm |
52.404 |
34.883 |
31.168 |
31.642 |
34.493 |
|
Yb |
54.392 |
38.508 |
32.354 |
30.697 |
31.047 |
|
Coif2 |
Mix. |
276.228 |
113.440 |
90.970 |
84.528 |
83.760 |
Er |
68.185 |
43.589 |
37.174 |
37.834 |
39.003 |
|
Tm |
51.813 |
34.415 |
30.914 |
31.251 |
31.713 |
|
Yb |
53.780 |
37.451 |
32.044 |
29.921 |
30.360 |
|
Coif4 |
Mix. |
277.145 |
107.641 |
86.738 |
83.806 |
87.135 |
Er |
68.182 |
42.591 |
37.072 |
37.435 |
41.578 |
|
Tm |
51.813 |
33.722 |
31.162 |
33.030 |
35.006 |
|
Yb |
53.752 |
36.947 |
32.133 |
31.387 |
33.073 |
|

Fig.5 The remained wavelet coefficients after compression and the extracted results
(wavelet basis: Symmlet 5, decomposition level: 7)
4.2 Resolved result by the proposed algorithm
In order to resolve the overlapping chromatogram by the proposed algorithm, both the
antigen (the multicomponent overlapping chromatogram) and the antibodies (the standard
chromatograms of each component) were compressed with Symmlet5 wavelet basis at
decomposition level 7. The number of data point is reduced from 52200 to 930. The solid
line in Figure 5 shows the compressed result of the overlapping chromatogram in Figure 2.
The dot lines show the resolved result by the IA. In order to see clearly, three different
regions are enlarged in Figure 6, in which (a) (b) (c) are corresponding to the data
points in the range of 140~190, 550~600, 660~690 respectively. It can be seen that the IA
can give a very good resolution of the compressed wavelet coefficients.
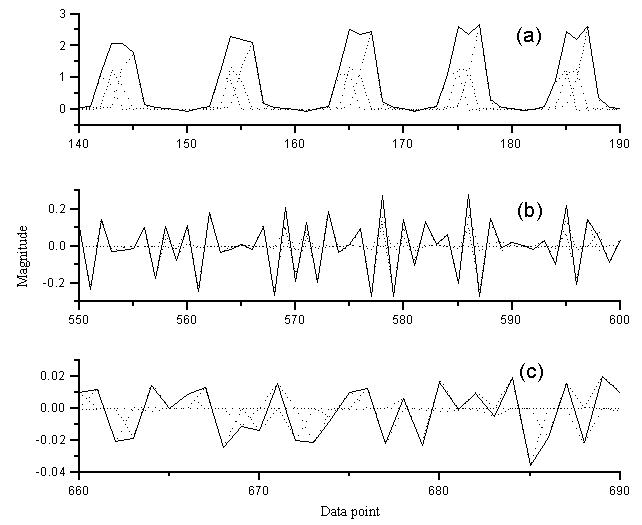
Fig.6 The enlargement of Figure 5
(a) 140~190 data point (b) 550~600 data point (c) 660~690 data point

Fig.7 The reconstructed 2-D chromatograms
(a) Er (b) Tm (c) Yb
The reconstructed
chromatograms from the resolved coefficients in Figure 5 are shown in Figure 7. (a) (b)
(c) are corresponding to the reconstructed chromatogram of each component respectively. In
comparison with the standard chromatogram of each component in Figure 3, it can be seen
that the overlapping chromatogram is well resolved and the chromatogram of each component
can be well obtained. The residual is shown in Figure 8, the intensity of the residual is
very small compared with that of the overlapping chromatograms or the reconstructed
chromatograms, which indicates that almost all the information contained in the
overlapping chromatogram was extracted. The little error is mainly caused by the
irreproducibility of the experiment.
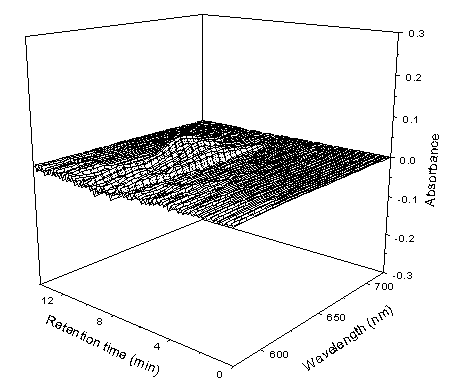
Fig.8 The reconstructed residual information
4.3 Comparison of the proposed algorithm with
immune algorithm and WT-IA
In our previous works, a WT-IA useing a two-dimensional wavelet compression algorithm was
proposed for the sake of improving the calculation speed. In order to investigate the
efficiency of the proposed algorithm, the consumed time and the residual after resolution
are compared with those of the IA and WT-IA, where the value of the residual is the
summation of every data point of the residual matrix as in Figure 8. The results are
listed in Table 3. It can be seen clearly that the speed of the proposed algorithm is 2.48
times faster than that of the IA, 2.16 times faster than that of WT-IA. The residual is
also smaller than that of IA and WT-IA.
Table 3 Comparison of conventional IA, WT-IA and the proposed algorithm*
No. Run |
Consumed time (s) |
Residual (×102) |
||||
Conv. IA |
WT-IA |
Proposed Algorithm |
Conv. IA |
WT-IA |
Proposed Algorithm |
|
1 |
18.56 |
16.37 |
7.47 |
2.1011 |
2.2326 |
2.0946 |
2 |
18.84 |
16.21 |
7.52 |
|||
3 |
18.84 |
16.36 |
7.58 |
|||
4 |
18.89 |
16.37 |
7.63 |
|||
5 |
18.51 |
16.37 |
7.58 |
|||
Aver. |
18.72 |
16.34 |
7.56 |
|||
* Program runs on Pentium(r)/233MHz/Memory 64M.
Table 4 Quantitative results by the proposed algorithm
Sample |
Added
Conc. |
Calculated
conc. |
Recovery |
|
1 |
Er |
0.2000 |
0.1977 |
98.85 |
Tm |
0.1999 |
0.1914 |
95.75 |
|
Yb |
0.2001 |
0.2070 |
103.45 |
|
2 |
Er |
0 |
0 |
0 |
Tm |
0.1499 |
0.1498 |
99.93 |
|
Yb |
0.2001 |
0.2046 |
102.25 |
|
3 |
Er |
0 |
0 |
0 |
Tm |
0.1999 |
0.1966 |
98.35 |
|
Yb |
0.2001 |
0.2041 |
102.00 |
|
4.4 Quantitative determination using the
proposed algorithm
In order to investigate the ability of the proposed algorithm for the quantitative
determination, the three samples listed in Table 2 were analyzed and the results were
listed in Table 4. It can be seen that all the recoveries are between 100± 5% with the
minimum being 95.72% and maximum being 103.44%. The results are satisfactory.
5. CONCLUSION
Based on the wavelet compression and immune algorithm, a fast algorithm for resolution of
2-D multicomponent overlapping chromatogram is proposed. By application of the method in
resolution and quantitative determination of multicomponent 2-D overlapping chromatograms,
it has been proven that this method is fast in calculation speed and accurate in
quantitative calculation. Therefore, the proposed algorithm may be an alternative
effective method for resolution of multicomponent 2-D overlapping chromatogram.
REFERENCES
[1] Maeder M. Anal. Chem., 1987, 59: 527.
[2] Schostack K J, Malinowski E R. Chemomtr. Intell. Lab. Syst., 1993, 20: 173.
[3] Shao X G, Cai W S, Sun P Y et al. Anal. Chem., 1997, 69: 1722.
[4] Shao X G, Cai W S, Sun P Y. Chemomtr. Intell. Lab. Syst., 1998, 43 (1,2): 147.
[5] Shao X G, Cai W S. Chem. J. Chin. Universities (Gaodeng Xuexiao Huaxue Xuebao), 1999,
20 (1): 42.
[6] Shao X G, Chen Z H, Lin X Q. Chemometr. Intell. Lab. Syst., 2000, 50 (1): 91.
[7] Shao X G, Chen Z H, Lin X Q. Fresenius' J. Anal. Chem., 2000, 366 (1): 10.
[8] Shao X G, Chen Z H, Lin X Q. Chin. J. Anal. Chem. (Fenxi Huaxue), 2000, 28 (2): 152.
[9] Sun L, Cai W S, Shao X G. Fresenius' J. Anal. Chem., 2001, 370 (1): 16.
[10] Shao X G, Sun L. Chem. J. Chin. Universities (Gaodeng Xuexiao Huaxue Xuebao), 2001,
22 (4): 552.
[11] Pelezer J, Szalma S. Chemical Reviews, 1991, 91: 1507.
[12] Alsberg B K, Nodland E, Kvalheim O M. J. Chemometr., 1993, 7: 61.
[13] Alsberg B K, Kvalheim O M. Chemometr. Intell. Lab. Syst., 1994, 24: 31.
[14] Alsberg B K, Kvalheim O M. Chemometr. Intell. Lab. Syst., 1994, 31: 43.
[15] Walczak B, Massart D L. Chemometr. Intell. Lab. Syst., 1997, 36: 81.
[16] Chau F T, Go J B, Shih T M et al. Appl. Spectrosc., 1997, 51: 649.
[17] Mallat S G. Trans. Amer. Math. Soc., 1989, 315: 69.
[18] Mallat S G. IEEE trans. Pattern Anal. Machine Intell., 1989, 11: 674.